Mycelium Dispersion from Fusarium oxysporum f. sp. dianthi Elicits a Reduction of Wilt Severity and Influences Phenolic Profiles of Carnation (Dianthus caryophyllus L.) Roots
Abstract
:1. Introduction
2. Results
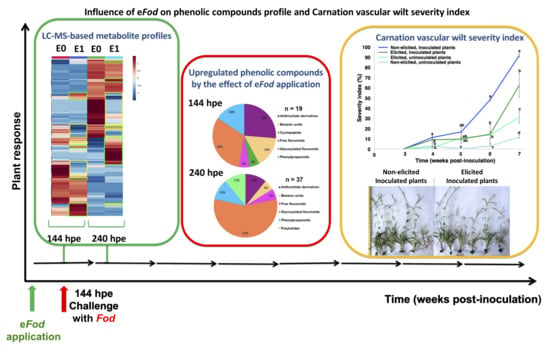
2.1. Phenotypic Response of Carnation to eFod
2.2. Effect of Elicitation on the Phenolic-Based Metabolite Profiles of ‘Mizuki’ and ‘Golem’ Cultivars
2.3. Effect of Elicitation on the Phenolic-Based Metabolite Profiles of Susceptible ‘Mizuki’ Cultivar Roots
2.4. Effect of Elicitation on the Phenolic-Based Metabolite Profiles of Resistant ‘Golem’ Cultivar Roots
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Preparation of the Biotic-Origin Elicitor (eFod)
4.3. In Vivo Test: Phenotypic and Metabolic Response of Carnation by Efod Application
4.3.1. Fod Inoculation
4.3.2. Evaluation of the Vascular Wilting Progress
4.3.3. Preparation of Raw Metabolite-Containing Extracts
4.3.4. LC-MS Analyses of Raw Extracts
4.4. Statistical Analyses
4.5. Annotation of Statistically Selected, Top-Ranked Metabolites by Elicitation Effect
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Benhamou, N. Elicitor-induced plant defence pathways. Trends Plant Sci. 1996, 1, 233–240. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Baenas, N.; García-Viguera, C.; Moreno, D.A. Elicitation: A tool for enriching the bioactive composition of foods. Molecules 2014, 19, 13541–13563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, D.R.; Ratsep, J.; Havis, N.D. Controlling crop diseases using induced resistance: Challenges for the future. J. Exp. Bot. 2013, 64, 1263–1280. [Google Scholar] [CrossRef]
- Burketova, L.; Trda, L.; Ott, P.G.; Valentova, O. Bio-based resistance inducers for sustainable plant protection against pathogens. Biotechnol. Adv. 2015, 33, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, N.; Nicole, M. Cell biology of plant immunization against microbial infection: The potential of induced resistance in controlling plant diseases. Plant Physiol. Biochem. 1999, 37, 703–719. [Google Scholar] [CrossRef]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef]
- Adrian, M.; Lucio, M.; Roullier-Gall, C.; Héloir, M.C.; Trouvelot, S.; Daire, X.; Kanawati, B.; Lemaître-Guillier, C.; Poinssot, B.; Gougeon, R.; et al. Metabolic fingerprint of PS3-induced resistance of grapevine leaves against Plasmopara viticola revealed differences in elicitor-triggered defenses. Front. Plant Sci. 2017, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mitra, A. Reinforcement of cell wall in roots of Lycopersicon esculentum through induction of phenolic compounds and lignin by elicitors. Physiol. Mol. Plant Pathol. 2007, 71, 201–209. [Google Scholar] [CrossRef]
- Chandra, S.; Chakraborty, N.; Dasgupta, A.; Sarkar, J.; Panda, K.; Acharya, K. Chitosan nanoparticles: A positive modulator of innate immune responses in plants. Sci. Rep. 2015, 5, 15195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, T.J.A. Variation in plant responsiveness to defense elicitors caused by genotype and environment. Front. Plant Sci. 2014, 5, 3–6. [Google Scholar] [CrossRef] [Green Version]
- TradeMap. Trade Statistics for International Business Development. Available online: https://www.trademap.org/ (accessed on 25 June 2021).
- Poli, A.; Bertetti, D.; Rapetti, S.; Gullino, M.L.; Garibaldi, A. Characterization and identification of Colombian isolates of Fusarium oxysporum f. sp. dianthi. J. Plant Pathol. 2013, 95, 255–263. [Google Scholar]
- Hegde, K.T.; Veeraghanti, K.S.; Kavita Hegde, C.T. Efficacy of bio-agents, botanicals and fungicides against Fusarium oxysporum f. sp. dianthi causing wilt of carnation. Int. J. Chem. Stud. IJCS 2017, 5, 139–142. [Google Scholar]
- Pérez Mora, W.; Melgarejo, L.M.; Ardila, H.D. Effectiveness of some resistance inducers for controlling carnation vascular wilting caused by Fusarium oxysporum f. sp. dianthi. Arch. Phytopathol. Plant Prot. 2020, 1–18. [Google Scholar] [CrossRef]
- Sánchez-Estrada, A.; Tiznado-Hernández, M.E.; Ojeda-Contreras, A.J.; Valenzuela-Quintanar, A.I.; Troncoso-Rojas, R. Induction of enzymes and phenolic compounds related to the natural defence response of netted melon fruit by a bio-elicitor. J. Phytopathol. 2009, 157, 24–32. [Google Scholar] [CrossRef]
- Troncoso-Rojas, R.; Sánchez-Estrada, A.; Carvallo, T.; González-León, A.; Ojeda-Contreras, J.; Aguilar-Valenzuela, A.; Tiznado-Hernández, M.E. A fungal elicitor enhances the resistance of tomato fruit to Fusarium oxysporum infection by activating the phenylpropanoid metabolic pathway. Phytoparasitica 2013, 41, 133–142. [Google Scholar] [CrossRef]
- Aldinary, A.M.; Morsy Abdelaziz, A.; Farrag, A.A.; Attia, M.S. Biocontrol of tomato Fusarium wilt disease by a new Moringa endophytic Aspergillus isolates. Mater. Today Proc. 2021. [Google Scholar] [CrossRef]
- De Borba, M.C.; de Freitas, M.B.; Stadnik, M.J. Ulvan enhances seedling emergence and reduces Fusarium wilt severity in common bean (Phaseolus vulgaris L.). Crop Prot. 2019, 118, 66–71. [Google Scholar] [CrossRef]
- Sathiyabama, M.; Charles, R.E. Fungal cell wall polymer based nanoparticles in protection of tomato plants from wilt disease caused by Fusarium oxysporum f. sp. lycopersici. Carbohydr. Polym. 2015, 133, 400–407. [Google Scholar] [CrossRef]
- Garcia-Brugger, A.; Lamotte, O.; Vandelle, E.; Bourque, S.; Lecourieux, D.; Poinssot, B.; Wendehenne, D.; Pugin, A. Early signaling events induced by elicitors of plant defenses. Mol. Plant Microbe Interact. 2006, 19, 711–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [Green Version]
- Lattanzio, V.; Kroon, P.A.; Quideau, S.; Treutter, D. Plant phenolics—secondary metabolites with diverse functions. In Recent Advances in Polyphenol Research; Daayf, F., Lattanzio, V., Eds.; Wiley-Blackwell: Oxford, UK, 2008; Volume 1, pp. 1–35. ISBN 9781444302400. [Google Scholar]
- Sterck, L.; Rombauts, S.; Vandepoele, K.; Rouzé, P.; Van de Peer, Y. How many genes are there in plants (... and why are they there)? Curr. Opin. Plant Biol. 2007, 10, 199–203. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Gamir, J.; Pastor, V.; Kaever, A.; Cerezo, M.; Flors, V. Targeting novel chemical and constitutive primed metabolites against Plectosphaerella cucumerina. Plant J. 2014, 78, 227–240. [Google Scholar] [CrossRef]
- Boba, A.; Kostyn, K.; Kostyn, A.; Wojtasik, W.; Dziadas, M.; Preisner, M.; Szopa, J.; Kulma, A. Methyl salicylate level increase in flax after Fusarium oxysporum infection is associated with phenylpropanoid pathway activation. Front. Plant Sci. 2017, 7, 1951. [Google Scholar] [CrossRef] [Green Version]
- Balmer, D.; De Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013, 74, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Bálintová, M.; Bruňáková, K.; Petijová, L.; Čellárová, E. Targeted metabolomic profiling reveals interspecific variation in the genus Hypericum in response to biotic elicitors. Plant Physiol. Biochem. 2019, 135, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Ardila, H.D.; Martínez, S.T.; Higuera, B.L. Levels of constitutive flavonoid biosynthetic enzymes in carnation (Dianthus caryophyllus L.) cultivars with differential response to Fusarium oxysporum f. sp. dianthi. Acta Physiol. Plant. 2013, 35, 1233–1245. [Google Scholar] [CrossRef]
- Sarrocco, S.; Falaschi, N.; Vergara, M.; Nicoletti, F.; Vannacci, G. Use of Fusarium oxysporum f. sp. dianthi transformed with marker genes to follow colonization of carnation roots. J. Plant Pathol. 2007, 89, 47–54. [Google Scholar]
- Ardila, H.D.; Baquero, B.; Martínez, S.T. Phenylalanine ammonium liase induction on carnation (Dianthus caryophyllus L.) by elicitors from Fusarium oxysporum f. sp. dianthi race 2 [text in spanish]. Rev. Colomb. Química 2007, 36, 151–167. [Google Scholar]
- Ardila, H.D.; Torres, A.M.; Martínez, S.T.; Higuera, B.L. Biochemical and molecular evidence for the role of class III peroxidases in the resistance of carnation (Dianthus caryophyllus L.) to Fusarium oxysporum f. sp. dianthi. Physiol. Mol. Plant Pathol. 2014, 85, 42–52. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Romero Rincón, A.E. Effect of the Application of Elicitors of Biotic Origin on the Biosynthesis of Flavonoids in Carnation (Dianthus caryophyllus L.) during the Interaction with Fusarium Oxysporum f. sp. Dianthi [Text in Spanish]; Universidad Nacional de Colombia: Bogotá, DC, Colombia, 2020; Available online: https://repositorio.unal.edu.co/handle/unal/78330 (accessed on 20 May 2021).
- Baayen, R.P.; Niemann, G.J. Correlations between accumulation of diantramides, dianthalexin and unknown compounds, and partial resistance to Fusarium oxysporum f. sp. dianthi in eleven carnation cultivars. J. Phytopathol. 1989, 126, 281–292. [Google Scholar] [CrossRef]
- Soto-Sedano, J.C.; Clavijo-Ortiz, M.J.; Filgueira-Duarte, J.J. Phenotypic evaluation of the resistance in F1 carnation populations to vascular wilt caused by Fusarium oxysporum f.sp. dianthi. Agron. Colomb. 2012, 30, 172–178. [Google Scholar]
- Ben-Yephet, Y.; Shtienberg, D. Effects of solar radiation and temperature on Fusarium wilt in carnation. Phytopathology 1994, 84, 1416–1421. [Google Scholar] [CrossRef]
- Walters, D.R.; Boyle, C. Induced resistance and allocation costs: What is the impact of pathogen challenge? Physiol. Mol. Plant Pathol. 2005, 66, 40–44. [Google Scholar] [CrossRef]
- Chakraborty, N.; Acharya, K. Ex vivo analyses of formulated bio-elicitors from a phytopathogen in the improvement of innate immunity in host. Arch. Phytopathol. Plant Prot. 2016, 49, 485–505. [Google Scholar] [CrossRef]
- Ross, A.F. Systemic acquired resistance induced by localized virus infections in plants. Virology 1961, 14, 340–358. [Google Scholar] [CrossRef]
- De Ascensao, A.R.F.D.C.; Dubery, I.A. Soluble and wall-bound phenolics and phenolic polymers in Musa acuminata roots exposed to elicitors from Fusarium oxysporum f. sp. cubense. Phytochemistry 2003, 63, 679–686. [Google Scholar] [CrossRef]
- Conrath, U.; Pieterse, C.M.; Mauch-Mani, B. Priming in plant-pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef] [Green Version]
- De-la-Cruz Chacón, I.; Riley-Saldaña, C.A.; González-Esquinca, A.R. Secondary metabolites during early development in plants. Phytochem. Rev. 2013, 12, 47–64. [Google Scholar] [CrossRef]
- Ponchet, M.; Favre-Bonvin, J.; Hauteville, M.; Ricci, P. Dianthramides (N-benzoyl and N-paracoumarylanthranilic acid derivatives) from elicited tissues of Dianthus caryophyllus. Phytochemistry 1988, 27, 725–730. [Google Scholar] [CrossRef]
- Lorenc-Kukuła, K.; Korobczak, A.; Aksamit-Stachurska, A.; Kostyń, K.; Łukaszewicz, M.; Szopa, J. Glucosyltransferase: The gene arrangement and enzyme function. Cell. Mol. Biol. Lett. 2004, 9, 935–946. [Google Scholar]
- Viladomat, F.; Bastida, J. General Overview of Plant Secondary Metabolism. In Plant Biology and Biotechnology; Bahadur, B., Rajam, M., Sahijram, L., Krishnamurthy, K., Eds.; Springer India: New Delhi, India, 2015; Volume 1, pp. 539–568. ISBN 9788132222866. [Google Scholar]
- Wink, M. Compartmentation of secondary metabolites and xenobiotics in plant vacuoles. Adv. Bot. Res. 1997, 25, 141–169. [Google Scholar]
- Beckman, C.H. Phenolic-storing cells: Keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 2000, 57, 101–110. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Takahama, U.; Hirota, S. Deglucosidation of quercetin glucosides to the aglycone and formation of antifungal agents by peroxidase-dependent oxidation of quercetin on browning of onion scales. Plant Cell Physiol. 2000, 41, 1021–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wink, M. Introduction: Biochemistry, physiology and ecological functions of secondary metabolites. In Biochemistry of Plant Secondary Metabolism; Wiley-Blackwell: Oxford, UK, 2010; Volume 40, pp. 1–19. ISBN 9781444320503. [Google Scholar]
- Chong, J.; Pierrel, M.A.; Atanassova, R.; Werck-Reichhart, D.; Fritig, B.; Saindrenan, P. Free and conjugated benzoic acid in tobacco plants and cell cultures. Induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol. 2001, 125, 318–328. [Google Scholar] [CrossRef] [Green Version]
- Higuera, B.L. Contribution to the Study of the Role of Phenolic Compounds in the Mechanisms of the Carnation Dianthus Caryophyllus L.-Fusarium Oxysporum f.sp. Dianthi [Text in Spanish]; Universidad Nacional de Colombia: Bogotá, DC, Colombia, 2001; Available online: https://repositorio.unal.edu.co/handle/unal (accessed on 20 May 2021).
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galeotti, F.; Barile, E.; Curir, P.; Dolci, M.; Lanzotti, V. Flavonoids from carnation (Dianthus caryophyllus) and their antifungal activity. Phytochem. Lett. 2008, 1, 44–48. [Google Scholar] [CrossRef]
- Abd-Elsalam, K.A.; Aly, I.N.; Abdel-Satar, M.A.; Khalil, M.S.; Verreet, J.A. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr. J. Biotechnol. 2003, 2, 96–103. [Google Scholar]
- Chiocchetti, A.; Bernardo, I.; Daboussi, M.J.; Garibaldi, A.; Gullino, M.L.; Langin, T.; Migheli, Q. Detection of Fusarium oxysporum f. sp. dianthi in carnation tissue by PCR amplification of transposon insertions. Phytopathology 1999, 89, 1169–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- León, K. Identification, Purification and Biochemical Characterization of 3 Proteases Secreted by Fusarium Oxysporum f. sp. Dianthi Race 2 In Vitro [Text in Spanish]; Universidad Nacional de Colombia: Bogotá, DC, Colombia, 2012; Available online: https://repositorio.unal.edu.co/handle/unal (accessed on 20 May 2021).
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Higuera, B.L.; De Gómez, V.M. Contribution of HPLC to the study of the defense mechanisms acting in carnation (Dianthus caryophyllus L.) Roots on infection with Fusarium oxysporum f. sp. dianthi. HRC J. High Resolut. Chromatogr. 1996, 19, 706–708. [Google Scholar] [CrossRef]
- Chiang, K.S.; Liu, H.I.; Bock, C.H. A discussion on disease severity index values. Part I: Warning on inherent errors and suggestions to maximise accuracy. Ann. Appl. Biol. 2017, 171, 139–154. [Google Scholar] [CrossRef]
- Monroy-Velandia, D.; Coy-Barrera, E. Effect of salt stress on growth and metabolite profiles of cape gooseberry (Physalis peruviana L.) along three growth stages. Molecules 2021, 26, 2756. [Google Scholar] [CrossRef] [PubMed]
- MetaboAnalyst 5.0. Available online: https://www.metaboanalyst.ca (accessed on 25 May 2021).
- Molecular Formula Finder. Available online: https://www.chemcalc.org/mf-finder (accessed on 26 May 2021).
- Dictionary of Natural Products. Available online: https://dnp.chemnetbase.com/ (accessed on 27 May 2021).
- KNApSAcK: A Comprehensive Species-Metabolite Relationship Database. Available online: http://www.knapsackfamily.com/knapsack_core/top.php (accessed on 27 May 2021).
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 27 May 2021).









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos-Rodríguez, J.; Coy-Barrera, E.; Ardila, H.D. Mycelium Dispersion from Fusarium oxysporum f. sp. dianthi Elicits a Reduction of Wilt Severity and Influences Phenolic Profiles of Carnation (Dianthus caryophyllus L.) Roots. Plants 2021, 10, 1447. https://doi.org/10.3390/plants10071447
Santos-Rodríguez J, Coy-Barrera E, Ardila HD. Mycelium Dispersion from Fusarium oxysporum f. sp. dianthi Elicits a Reduction of Wilt Severity and Influences Phenolic Profiles of Carnation (Dianthus caryophyllus L.) Roots. Plants. 2021; 10(7):1447. https://doi.org/10.3390/plants10071447
Chicago/Turabian StyleSantos-Rodríguez, Janneth, Ericsson Coy-Barrera, and Harold Duban Ardila. 2021. "Mycelium Dispersion from Fusarium oxysporum f. sp. dianthi Elicits a Reduction of Wilt Severity and Influences Phenolic Profiles of Carnation (Dianthus caryophyllus L.) Roots" Plants 10, no. 7: 1447. https://doi.org/10.3390/plants10071447