Sign up here to subscribe to the Grower2grower Ezine. Every two weeks you will receive new articles, specific to the protected cropping industry, informing you of industry news and events straight to your inbox.
Sep 2023
F. fulva have now evolved worldwide to overcome both the Cf-9C and Cf-9B resistance genes

Sequential breakdown of the most commonly deployed leaf mould resistance locus in tomato by Fulvia fulva (formerly Cladosporium fulvum)

Compound leaves from tomato cultivars that are susceptible (left) or resistant (right) to leaf mould disease. Photos taken at two weeks post-inoculation. Credit: Carl Mesarich, Massey University.
Leaf mould, caused by the fungus Fulvia fulva (formerly Cladosporium fulvum), is a devastating disease of tomato plants grown in humid environments such as greenhouses. Left untreated, the disease can result in severe defoliation and yield losses.
Upon landing on the lower leaf surface, spores of F. fulva germinate to produce runner hyphae that enter the leaf apoplast through natural openings called stomata. Once inside the apoplast, the fungus delivers a collection of virulence factors, termed effector proteins, to promote colonization and disease. In susceptible tomato cultivars, the fungus eventually reemerges from stomata to produce masses of spores that then go on to cause secondary infections.
Tomato cultivars that are resistant to leaf mould disease possess one or more disease resistance genes. These genes encode cell surface-localized resistance proteins that recognize specific effector proteins of F. fulva in the apoplast. Following the recognition of an effector protein, defence responses are initiated that render the tomato cultivar resistant.
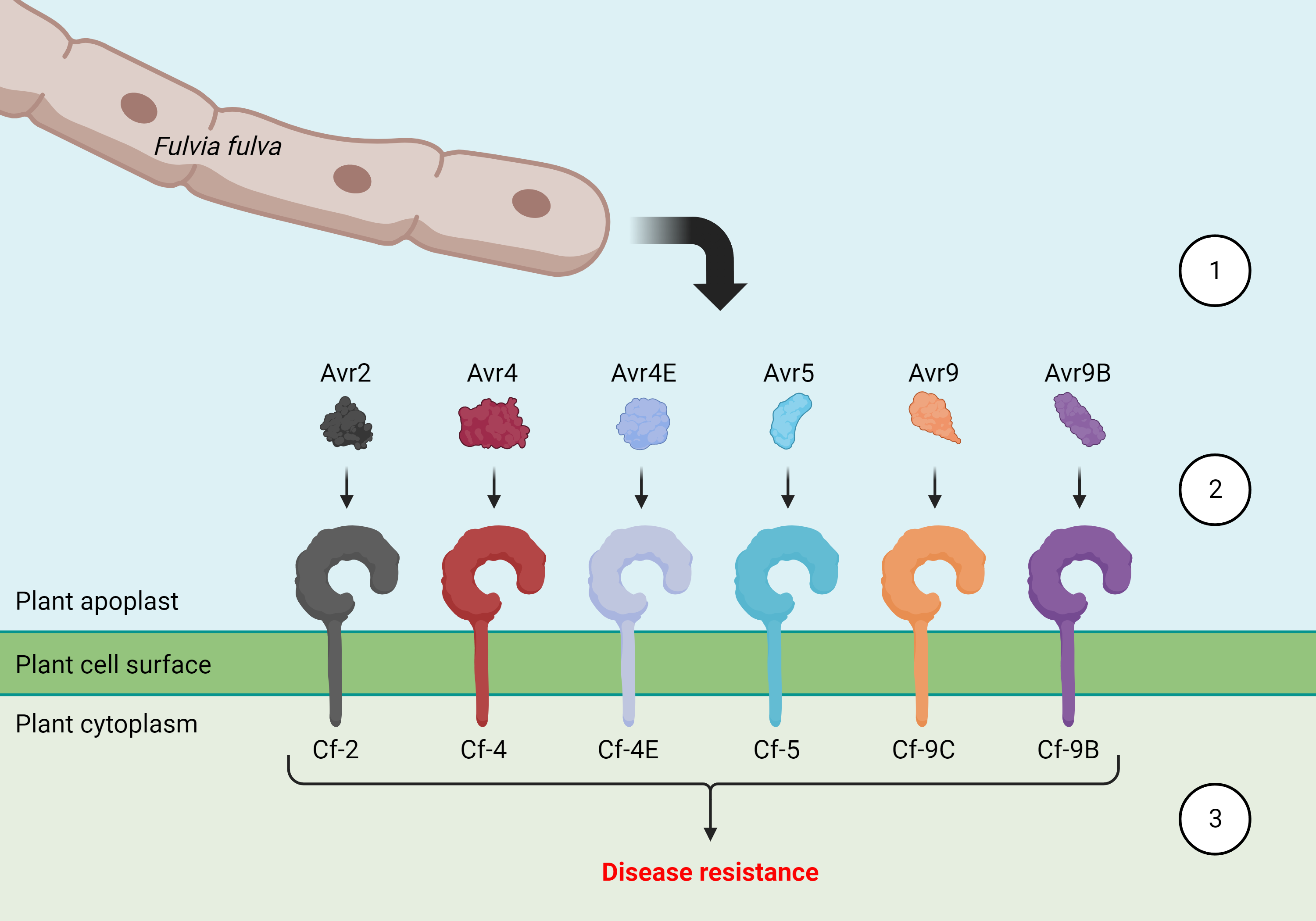
Molecular interactions underpinning resistance to tomato leaf mould disease. The tomato leaf mould fungus, Fulvia fulva, delivers a collection of virulence factors, termed effector proteins, into the apoplast of tomato leaves to promote colonization and disease. Amongst these are the effector proteins Avr2, Avr4, Avr4E, Avr5, Avr9, and Avr9B (step 1). Tomato cultivars that are resistant to leaf mould disease, possess one or more disease resistance genes. These genes encode cell surface-localized resistance proteins that recognize specific effector proteins of F. fulva in the apoplast. For example, the Cf-9C resistance protein recognizes the Avr9 effector protein, while the Cf-9B resistance protein recognizes the Avr9B effector protein (step 2). Following recognition of an effector protein by a resistance protein, defence responses are initiated that render the tomato cultivar resistant to leaf mould disease (step 3). Image created with www.BioRender.com
Notably, most resistant tomato cultivars that are commercially available are designated Ff:A–E, which means that they carry the Cf-9 resistance locus. This locus is made up of five genes, two of which encode resistance proteins that provide protection against leaf mould disease. These are Cf-9C, which recognizes the Avr9 effector protein and provides resistance during all stages of plant growth, and Cf-9B, which recognizes the Avr9B effector protein and provides resistance to mature plants only.
Shortly after the deployment of the Cf-9 resistance locus in tomato cultivars during the 1970s, F. fulva strains began to emerge that could overcome the Cf-9C resistance gene through deletion (loss) of the corresponding Avr9 effector gene. Despite this, the Cf-9B resistance gene proved to be much more durable, providing growers with protection against leaf mould disease during the important flowering and fruiting period. More recently, however, growers from around the world have reported an increase in the incidence of F. fulva strains capable of infecting mature Cf-9 tomato plants. This indicates that strains of F. fulva have now evolved worldwide to overcome both the Cf-9C and Cf-9B resistance genes.
In an article recently posted to the preprint server for biology, bioRxiv (de la Rosa and Schol et al., 2023), researchers from Massey University in New Zealand, led by the research group of Dr. Carl Mesarich, and in conjunction with collaborators from Wageningen University and Research in the Netherlands, Setsunan University in Japan, as well as the New Zealand Institute for Plant and Food Research, set out to investigate the molecular mechanism underpinning the breakdown of the Cf-9 resistance locus by F. fulva. By sequencing the genome of a New Zealand F. fulva strain that had overcome the Cf-9 locus in the 1980s (the first strain reported to do so) and comparing it to the genome of a reference strain that had not overcome the Cf-9 locus, they were able to identify the Avr9B gene and show that it had been deleted in the New Zealand strain.
Following on from this, the authors identified eight additional Cf-9B-breaking mutations in Avr9B across F. fulva strains collected from the Netherlands, Germany, France, Tanzania, China, and Japan. Strikingly, in F. fulva strains recently collected from Cf-9 plants, there was a strict correlation between Avr9 deletion and resistance-breaking mutations in Avr9B. In contrast, in older strains, Avr9 had been deleted but no mutations were observed in Avr9B. This suggests that the Cf-9 resistance locus has been sequentially broken down through the evolution of F. fulva strains over time, with Cf-9C circumvented prior to Cf-9B. As Cf-9B only provides resistance to mature plants, it is suspected that significant selection pressure was first placed on F. fulva to overcome Cf-9C-mediated resistance in young plants. Then, once Cf-9C-mediated resistance had been overcome through deletion of the Avr9 gene, there was increasing selection pressure exerted on F. fulva to overcome Cf-9B-mediated resistance in mature plants, resulting in the loss or mutation of the Avr9B gene.
In terms of resistance to leaf mould disease, what does this mean for the continued use of tomato cultivars with the Cf-9 locus? As F. fulva strains have overcome both Cf-9B and Cf-9C worldwide, the authors of the study suggest that the Cf-9 resistance locus now has limited value for controlling leaf mould disease in commercial tomato production. While other tomato cultivars with resistance to leaf mould disease are available, these typically only contain one resistance gene that has already been overcome by F. fulva. Certainly, strains of F. fulva now exist that can break down one or more of the well-studied Cf-2, Cf-4, Cf-4E, and Cf-5 resistance genes through mutation or deletion of the corresponding Avr2, Avr4, Avr4E, and Avr5 effector genes, respectively. This is often in addition to gene deletion or mutation events that have allowed these strains to overcome resistance provided by the Cf-9 locus.
So, what needs to be done now? Moving forward, multiple resistance genes that have not yet been overcome by F. fulva, and that simultaneously provide protection to leaf mould disease during all stages of plant growth, need to be stacked into tomato cultivars. Crucially, progress has been made towards this objective, with wild tomato species recently identified that likely carry one or more of 10 new resistance genes active against leaf mould disease (Mesarich et al., 2018). It is hoped that in stacking these resistance genes, durable resistance can be achieved to support future commercial tomato production.
For more information about the study posted on bioRxiv, please click the link below or contact the corresponding author, Dr. Carl Mesarich (c.mesarich@massey.ac.nz).
de la Rosa S, Schol CR, Peregrina ÁR, Winter DJ, Hilgers AM, Maeda K, Iida Y, Tarallo M, Jia R, Beenen HG, Rocafort M, de Wit PJGM, Bowen JK, Bradshaw RE, Joosten MHAJ, Bai Y, Mesarich CH. (2023). Sequential breakdown of the complex Cf-9 leaf mould resistance locus in tomato by Fulvia fulva. bioRxiv, 2023.08.27.554972. DOI: https://doi.org/10.1101/2023.08.27.554972. Please note: this article is a preprint and has not yet been certified by peer review.
Mesarich CH, Ökmen B, Rovenich H, Griffiths SA, Wang C, Karimi Jashni M, Mihajlovski A, Collemare J, Hunziker L, Deng CH, van der Burgt A, Beenen HG, Templeton MD, Bradshaw RE, de Wit PJGM (2018). Specific hypersensitive response-associated recognition of new apoplastic effectors from Cladosporium fulvum in wild tomato. Molecular Plant-Microbe Interactions, 31(1): 145–62. DOI: https://doi.org/10.1094/MPMI-05-17-0114-FI.
CLASSIFIED
Photo
Gallery
Subscribe to our E-Zine
More
From This Category

A crop on the cusp of ruin by leaf mould and the return of psyllid yellows.

Signify foresees a bright future for vertical farming, driven by automation and diversification.

Revitalise your orchard – the use of Sili-Fert P for next-level apple quality

More efficient cultivation with controlled raising under LED lighting
GrowUp: Serving the UK the best in fresh food













































