- 1Department of Biotechnology, College of Life Science and Technology, Institute of Resource Biology and Biotechnology, Huazhong University of Science and Technology, Wuhan, China
- 2Key Laboratory of Molecular Biophysics Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
- 3Hubei Engineering Research Center for Edible and Medicinal Resources, Wuhan, China
Taxol is a rare secondary metabolite that accumulates considerably in Taxus species under salicylic acid (SA) and methyl jasmonate treatment. However, the molecular mechanism of its accumulation remains unclear. We investigated TcWRKY33, a nuclear-localized group I WRKY transcription factor, as an SA-responsive regulator of taxol biosynthesis. Overexpression and RNA interference of TcWRKY33 confirmed that TcWRKY33 regulates the expression of most taxol biosynthesis genes, especially 10-deacetylbaccatin III-10-O-acetyltransferase (DBAT) and taxadiene synthase (TASY), which were considered as key enzymes in taxol biosynthesis. Transient overexpression of TcWRKY33 in Taxus chinensis leaves resulted in increased taxol and 10-deacetylbaccatin accumulation by 1.20 and 2.16 times compared with the control, respectively. Furthermore, TcWRKY33, DBAT, and TASY were confirmed to respond positively to SA signals. These results suggested that TcWRKY33 was the missing component of taxol biosynthesis that responds to SA. The sequence analysis identified two W-box motifs in the promoter of DBAT but not in the TASY. Yeast one-hybrid and dual-luciferase activity assays confirmed that TcWRKY33 can bind to the two W-boxes in the promoter of DBAT, upregulating its expression level. Hence, DBAT is a direct target of TcWRKY33. Furthermore, TcERF15, encoding a TASY activator, also contains two W-boxes in its promoter. Yeast one-hybrid and dual-luciferase activity assays further confirmed that TcWRKY33 can upregulate TASY expression through the activation of TcERF15. In summary, TcWRKY33 transmits SA signals and positively regulates taxol biosynthesis genes in two ways: directly and through the activation of other activators. Therefore, TcWRKY33 is an excellent candidate for genetically engineering regulation of taxol biosynthesis in Taxus plants.
Introduction
Taxol, also known as paclitaxel, is a specialized metabolite originally isolated from Taxus species that functions as an anticancer drug (Wani et al., 1971; Schiff et al., 1979). Similar to other secondary plant metabolites, such as artemisinin and vinblastine, taxol is very rare in vivo (Kwak et al., 1995; White, 2008; Abdul Rahim et al., 2018). More than 20 enzymes participate in the taxol biosynthesis pathway, making taxol biosynthesis more complex than the biosynthesis of artemisinin and camptothecin (Croteau et al., 2006). Thus, studies of the regulatory mechanism of taxol biosynthesis are beset with difficulties, resulting in inconsistency between commercial supply and the clinical requirements for taxol.
Salicylic acid (SA) is an important endogenous hormone in plants. It is involved in the regulation of the biosynthesis of secondary metabolites including terpenoids, alkaloids, and flavonoids (Kang et al., 2004; Tounekti et al., 2013). In suspension cultures of Taxus chinensis var. mairei, the addition of 20 mg/L of SA induces taxol biosynthesis (Wang et al., 2007). Miao et al. (2000) reported that the addition of 0.1 mg/L of SA can enhance the production of taxol by up to 3-fold. SA also increases the concentrations of 10-deacetylbaccatin (10-DAB) and baccatin III (Miao et al., 2000). Furthermore, many transcription factors function by responding to SA signals. PtrWRKY73, OsWRKY77, and BcWRKY46 respond to SA signals during defense responses (Wang et al., 2012; Lan et al., 2013; Duan et al., 2015). However, SA-induced transcription factors that regulate taxol biosynthesis remain to be discovered.
Only a few transcription factors are known to regulate the expression of taxol biosynthesis genes (Zhang et al., 2018b; Cui et al., 2019). Li et al. (2013) reported that the group I WRKY transcription factor TcWRKY1 can bind two W-boxes in the promotor of 10-deacetylbaccatin III-10-O-acetyltransferase (DBAT) to upregulate its expression in T. chinensis. Lenka et al. (2015) identified three bHLH factors, TcJAMYC1/2/4, showing potential for downregulating several taxol biosynthesis genes, such as phenylpropanoyltransferase (BAPT), by binding to G- and E-boxes. In addition, TcJAMYC2 and TcJAMYC4 induce reporter gene expression under the control of taxadiene 5-alpha hydroxylase (T5H) and taxadiene synthase (TASY) promoters, respectively (Lenka et al., 2015). Moreover, two ERF factors (TcERF12 and TcERF15) and one MYC2 factor (TcMYC2a) strongly regulate the TASY gene by interacting with GCC- and G-like-boxes, respectively (Zhang et al., 2015, 2018b). All these factors are focused on the methyl jasmonate (MJ) signaling transduction pathway and may therefore effectively regulate taxol biosynthesis. Currently, no transcription factors responding to SA have been reported. Hence, this regulatory mechanism should be investigated.
WRKY transcription factors belong to one of the largest transcription factor families in plants (Eulgem et al., 2000; Robatzek and Somssich, 2001). Several WRKYs respond to SA treatment and play comprehensive and important roles in diverse physiological processes. For example, PtrWRKY73, a group I WRKY transcription factor, responds to SA treatment and plays a positive role in plant resistance to biotrophic pathogens (Duan et al., 2015). The SA-induced OsWRKY77 transcription factor functions as a positive regulator of defense/pathogenesis-related genes and makes transgenic plants highly resistant to infection by pathogens (Lan et al., 2013). In Chinese wild Vitis quinquangularis, VqWRKY52 plays essential roles in the SA-dependent signal transduction pathway and is capable of enhancing hypersensitive response cell death triggered by microbial pathogens (Wang et al., 2017).
In this study, we evaluated the potential functions of the group I WRKY transcription factor TcWRKY33 in the regulation of taxol biosynthesis. We confirmed its induction by SA treatment and performed overexpression and RNA interference experiments to determine its effect on the content of taxol and related taxanes. Expression levels of taxol biosynthesis genes were determined by using quantitative real-time PCR (qRT-PCR). We subsequently investigated the molecular regulation mechanism of TcWRKY33 through yeast one-hybrid assays, dual-luciferase reporter systems, and evaluation of expression patterns. Our results broaden our understanding of the positive roles of TcWRKY33 transcription factors in regulating the biosynthesis of taxol in plants.
Materials and Methods
Plant Hormone Treatment
Taxus chinensis cell line #48 was maintained in modified Gamborg's B5 medium (62# medium) (Zhang et al., 2018a). Six-gram long-term sub-cultured cells of T. chinensis were collected into 62# liquid medium at 125 rpm for 48 h dark period. Then, 0.1 mmol/L of MeJA or 2.5 mmol/L of SA was added into the medium, respectively. The samples were taken after 0, 1, 3, and 6 h and ground in liquid nitrogen.
Total RNA Isolation, cDNA Synthesis, and Cloning of TcWRKY33 Gene
Fresh leaves of T. chinensis were collected from the Western Nursery Garden of Huazhong University of Science and Technology, Vilnius, Lithuania. Harvest samples were ground in liquid nitrogen and used for the isolation of total RNA. Total RNA isolation was conducted by using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China, Cat: DP441), and the first chain of cDNA was transcribed by using the RevertAid First Strand cDNA Synthesis Kit (Thermo, USA, Cat: K1621).
The sequence of TcWRKY33 gene was obtained from the multi-omics analysis data. It was found to harbor two WRKY domains in its deduced amino acid sequences, and its full-length was amplified by using reverse transcription-PCR. The primers named TcWRKY33-1F/R are shown in Supplementary Table 1. After amplifying with the template of T. chinensis cDNA, PCR products were T/A cloned into the pMD18-T vector (TaKaRa, Dalian, China) and sequenced.
Sequence Alignment and Phylogenetic Analyzing of TcWRKY33
BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) was used for the homology search from the Swiss-Prot database. ClustalW online (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html) at the default setting was used to align and evaluate the percentage identity of TcWRKY33 with known WRKYs. The alignment results were sent to ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) for coloring (Robert and Gouet, 2014). The phylogenetic analysis was performed with a neighbor-joining method by using 1,000 bootstrap resamplings in MEGA 7.0.
Subcellular Localization
The full-length TcWRKY33 Open Reading Frame (ORF) without termination codon was amplified using the primers TcWRKY33-2F/R in Supplementary Table 1 and then inserted into pCAMBIA1300-35S-sGFP with SacI and BamHI restriction sites, and then recombinant vector was transformed into GV3101. The onion epidermal cells with a dimension of 1 × 1 cm were infected by positive clones and controls (pCAMBIA1300-35S-sGFP) in 1/2 MS liquid medium at 25°C in dark for 2 days. The infected onion epidermal cells were then plated on the MS medium for 2 days at 25°C in darkness. The GFP fluorescence was observed by using the laser scanning confocal microscope FV1000 (OLYMPUS, Japan).
Cloning, Activity Validation, and Promoter Analysis of the 5′-Flanking Sequence of TcERF15 Gene
First, the Coding DNA Sequence (CDS) of TcERF15 of T. chinensis was aligned with Taxus baccata genome sequences, suggesting that it was highly conserved in both Taxus spp. In addition, the 5′-flanking sequence of this gene of T. baccata was obtained. To obtain and validate the similarity of flanking sequences of this gene in T. chinensis, the forward primer was designed according to the homolog sequence of T. baccata, while the reverse primer was designed according to the ORF sequence in T. chinensis (Supplementary Table 1). After amplifying with the template of T. chinensis genomic DNA, PCR products were T/A cloned and sequenced. Only those that contain 5′-UTR Untranslated Regions and partial ORF sequence of TcERF15 were considered as the 5′-flanking sequence in T. chinensis.
Then, the fragment of TcERF15 was inserted into pBI121 with HindIII and BamHI restriction sites, so that the β-glucuronidase (GUS) reporter was under the control of the promoter of TcERF15. The construct was transformed into onion epidermis cells mediated by GV3101 affection (Zhang et al., 2018b). To test its promoter activity, GUS staining was conducted according to our previous report (Li et al., 2013).
Then, the 5′-flanking sequence was analyzed to locate basic cis-elements and W-boxes by using PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and Nsite (http://www.softberry.com/berry.phtml?topic=nsite&group=programs&subgroup=promoter).
Yeast One-Hybrid
For prey vectors, TcWRKY33 was ligated into pGADT7-Rec2 with EcoRI and BamHI restriction sites, resulting in a TcWRKY33::pGADT7-Rec2 recombinant vector (Supplementary Table 1). For bait vectors, sense, and antisense of 6-bp fragments containing W-boxes from promoters that were quadrupled with cohesive ends of EcoRI and SacI were artificially synthesized, then the double strand generated by annealing was linked with digested pHIS2.1 vector, resulting in a series of bait vectors (Supplementary Table 1). Finally, each bait vector was co-transformed with the prey vector into the yeast strain Y187, and then they were plated onto the SD/Leu-/Trp-/His-deficient medium with 20/40 mM 3-AT for 3–5 days. Each bait vector co-transformed with empty pGADT7-Rec2 was used as the negative control.
Construction of Overexpression and RNAi Vectors
The full-length of TcWRKY33 was amplified from TcWRKY33::pMD18-T vector using the specific primers TcWRKY33-3F/R in Supplementary Table 1 and inserted into the vector with NcoI restriction site to generate the overexpression vector TcWRKY33::pCAMBIA3302. For the RNAi vector, a 376 bp 3′-CDS fragment of TcWRKY33 was forward and reversely inserted into the LH-FAD2-1390RNAi vector so that the two fragments and the FAD-RNAi fragment would generate a stem-loop in cells (Supplementary Table 1). Then, the recombinant vectors were transformed into LBA4404, and LBA4404 containing empty pCAMBIA3302 or LH-FAD2-1390RNAi vector was used as the control, respectively.
Transient Transformation of TcWRKY33 in T. chinensis Leaves
For transient transformations of Taxus leaves, the single colonies of LBA4404 containing empty vectors, TcWRKY33::pCAMBIA3302 and TcWRKY33::LH-FAD2-1390RNAi, were cultivated in 20 ml of LB medium (LB) until OD600 = 0.6–0.8, and then they were collected and resuspended with 20 ml of half-strength MS liquid medium containing 200 μM of acetosyringone. The fresh leaves were gathered from blade tips on the 10-year-old T. chinensis potted plants. After surface disinfection, small incisions were made with a sterile scalpel in the inner surface of the leaves to facilitate Agrobacterium infection. Then, the injured leaves were cultured in a shaker at 125 rpm for 12 h in the dark at 28°C. After that, the infected T. chinensis leaves were plated on the 1/2 MS solid medium for 24 h at 28°C in darkness. Finally, the part of leaves was placed into the liquid nitrogen and then stored in the fridge at −80°C for the following experiments, and the others were dipped into sterile water shaking at 120 rpm for 6 days to test the contents of the taxol and 10-DAB.
Quantitative Real-Time PCR and High-Performance Liquid Chromatography
Quantitative real-time PCR was conducted as previously described (Zhang et al., 2015), and three fully independent biological replicates were obtained for the statistical analysis (Supplementary Table 1). Each experiment was conducted three times, and we used Student's t-test. The treated leaves were dried at 60°C for 24 h and crushed to 40 meshes, 0.1 g of powder was first weighed accurately and put into a 10-ml centrifuge tube, and then 3 ml of methanol were added. The extraction procedure was conducted by using an ultrasonic machine (Kunshan KQ-250E, China) for 30 min, centrifuged for 10 min at 8,000 r/min, and repeated three times, and the upper phase was collected and dried using N-Evap (Autoscience MTN-2800D, China). The taxane samples were diluted by high-performance liquid chromatography (HPLC)-grade methanol and then passed through membrane filters (0.22 μm) before HPLC detection. Each experiment was conducted in triplicate. The extraction compounds were analyzed using the Agilent 1290 HPLC system combined with a phenyl-C18 column (250 × 4.6 mm, 4.6 μm). Chromatographic separation was performed by the gradient elution process of mobile phase water (A) and acetonitrile (B) as follows: 0–10 min, 5–40% B; 10–15 min, 40–50% B; 15–30 min, 50–80% B; 30–35 min, 80–95% B. The flow rate was 1.0 ml/min with 20 μl of injection volume. In addition, the column temperature and the detection wavelength were kept at 30°C and 227 nm, respectively.
Effector–Reporter Assays
For dual-luciferase activity assay, the original 383 bp DBAT promoter/715 bp TcERF15 promoter and two progressive deletions removing the Wa-box/W1-box and Wb-box/W2-box were cloned with restriction sites of HindIII and BamHI and inserted into the pGreenII-0800-Luc vector to generate the reporter vector (Supplementary Table 1). Then, TcWRKY33::pCAMBIA3302 acted as the effector vector. Each reporter and effector were transiently expressed in Nicotiana benthamiana leaves via Agrobacterium tumefaciens strain GV3101. The infiltrated plants were grown in a greenhouse for 24 h in darkness and then grown under natural conditions for 2 days, and the luciferase activities were measured with a dual-luciferase reporter assay kit (Vazyme).
Results
TcWRKY33 Is an SA-Responsive Factor
Salicylic acid and MJ enhance the production of taxol (Yukimune et al., 1996; Miao et al., 2000). WRKY transcription factors are important SA and MJ signaling regulators, and DBAT and TASY respond to SA and MJ signals (Zhang et al., 2017). Analyzing the hormonal response patterns of TcWRKY33 will therefore enrich our understanding of the regulatory network of taxol biosynthesis.
Phytohormone treatment was conducted in T. chinensis cells and we measured the expression of TcWRKY33 by using qRT-PCR. Expression of TcWRKY33 was slightly higher at 1 and 3 h after SA treatment compared with that before treatment but increased by 3.24 times at 6 h after the induction of SA. However, TcWRKY33 expression showed no obvious difference after MJ treatment. Hence, TcWRKY33 responded significantly to the SA signal but not to MJ (Figure 1A). The response of TcWRKY33 to SA was not immediate; accumulation of TcWRKY33 transcripts after the induction of SA took time.
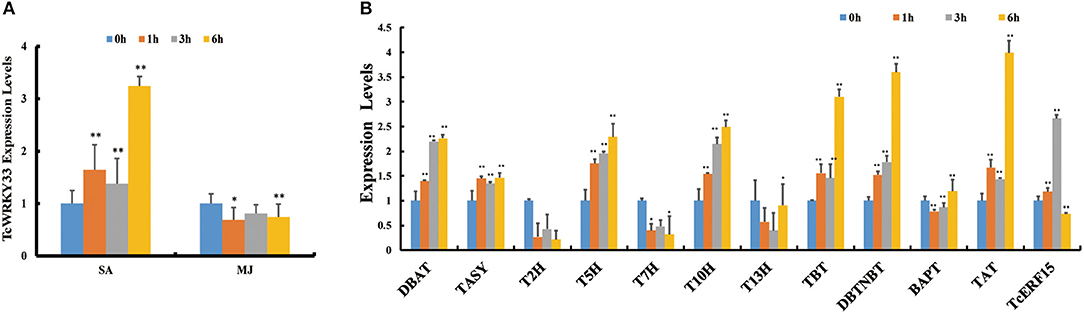
Figure 1. Induction of TcWRKY33 by salicylic acid (SA) and methyl jasmonate (MJ) and the expression of taxol biosynthesis genes and TcERF15 in SA treatment. (A) Relative expression of TcWRKY33 in response to SA and MJ. (B) Relative expression of TcERF15 and taxol biosynthesis genes in response to SA. Cells were harvested at different times (0, 1, 3, and 6 h) after treatment. Actin was used as an endogenous reference. The statistics method of p-value was Student's t-test. Asterisks indicate the significance, *0.01 < p < 0.05, and **p < 0.01.
In addition to TcWRKY33, many taxol biosynthesis genes also responded to SA signals. Expression levels of DBAT, TASY, T5H, 5-alpha-taxadienol-10-beta-hydroxylase (T10H), taxane 2-alpha-O-benzoyltransferase, 3′-N-debenzoyltaxol N-benzoyltransferase (DBTNBT), and taxadienol acetyltransferase (TAT) were increased by 2.26-, 1.47-, 2.29-, 2.49-, 3.10-, 3.60-, and 3.99-folds, respectively, at 6 h after SA treatment compared with control (Figure 1B), indicating that these taxol biosynthesis genes respond positively to SA signals.
TcWRKY33 Is a Nuclear-Localized Group I WRKY Transcription Factor
An online Blast search of the SWISS-Prot database showed that TcWRKY33 has the most similarity to the WRKY transcription factors, SUSIBA2 and AtWRKY20, and might thus function as an activator (Sun et al., 2003; Nagata et al., 2012). Sequence alignment revealed that the putative TcWRKY33 contained two conserved WRKY domains in the N-terminus and C-terminus, followed by a Cys2/His2-type zinc-finger motif; it, therefore, represents a member of the group I WRKY transcription factors (Figure 2A). The TcWRKY33 protein also contained an LSPLL motif, which is present in many transcription factors and cofactors and mediates interactions that activate or repress transcription (Plevin et al., 2005). The phylogenic analysis indicated that the TcWRKY33 protein was closely related to the group I WRKY family members, sharing the highest similarity with AtWRKY20 (Figure 2B).
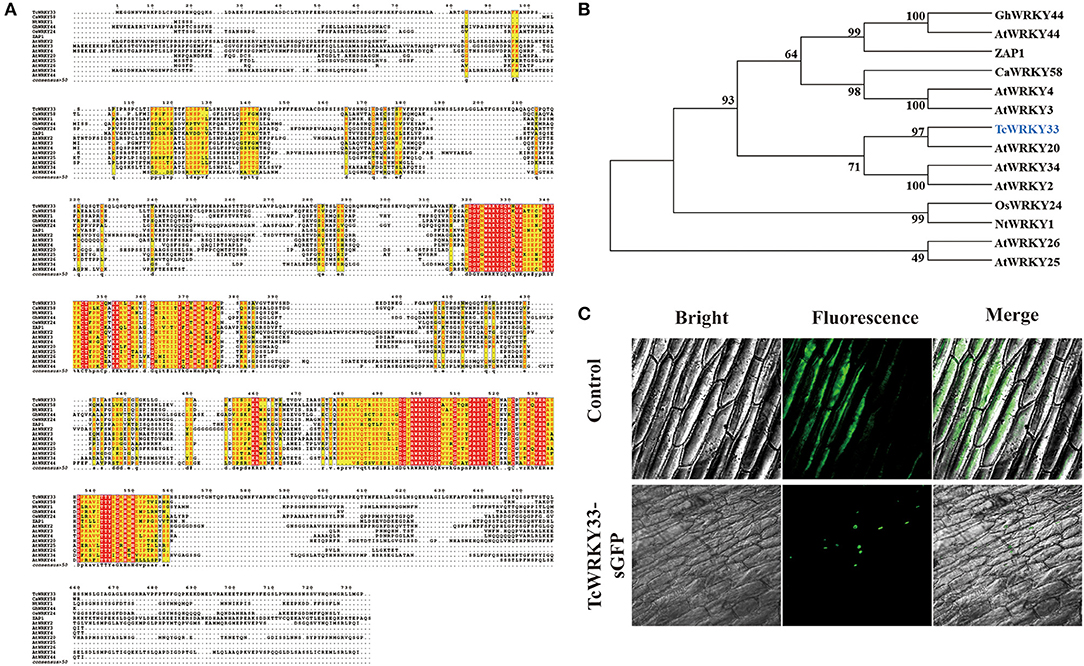
Figure 2. Sequence alignment, phylogenetic analysis, and subcellular localization of TcWRKY33. (A) Amino acid sequence alignment of TcWRKY33 and other WRKYs in Arabidopsis thaliana, Capsicum annuum, Nicotiana tabacum, and Oryza Sativa. (B) Phylogenetic analysis of TcWRKY33 and other WRKYs. TcWRKY33 was signed in blue. (C) The fused protein of TcWRKY33 and GFP was transformed into onion epidermal cells. GFP fluorescence was observed 2 days after infecting by using a laser scanning confocal microscope FV1000 (OLYMPUS, Japan) with 10 × 20 magnification.
Subcellular localization in onion epidermal cells to verify the site of TcWRKY33 activity revealed that TcWRKY33 is a nuclear-localized protein (Figure 2C). Most transcription factors function in the nucleus; in particular, WRKY factors are DNA-binding proteins, suggesting their localization in the nucleus (Ciolkowski et al., 2008).
TcWRKY33 Upregulates Seven Genes Involved in Taxol Biosynthesis
The biosynthesis of taxol is a complex process and it involves many enzymes (Figure 3A). To identify the function of TcWRKY33 in regulating the expression of taxol biosynthesis genes, we transiently overexpressed TcWRKY33 in Taxus leaves. Expression levels of taxol biosynthesis genes were examined by using qRT-PCR. The expression of TcWRKY33 in transformed leaves was 5.72 times greater than that in the control (Figure 3B). Transcriptional levels of DBAT, TASY, 2-alpha-hydroxylase (T2H), T5H, T10H, TAT, and DBTNBT were 3.85-, 2.27-, 2.81-, 2.01-, 1.74-, 2.84-, and 4.19-fold higher in leaves overexpressing TcWRKY33 than in controls (Figure 3B). In contrast, the BAPT expression was 0.57 times that of controls (Figure 3B).
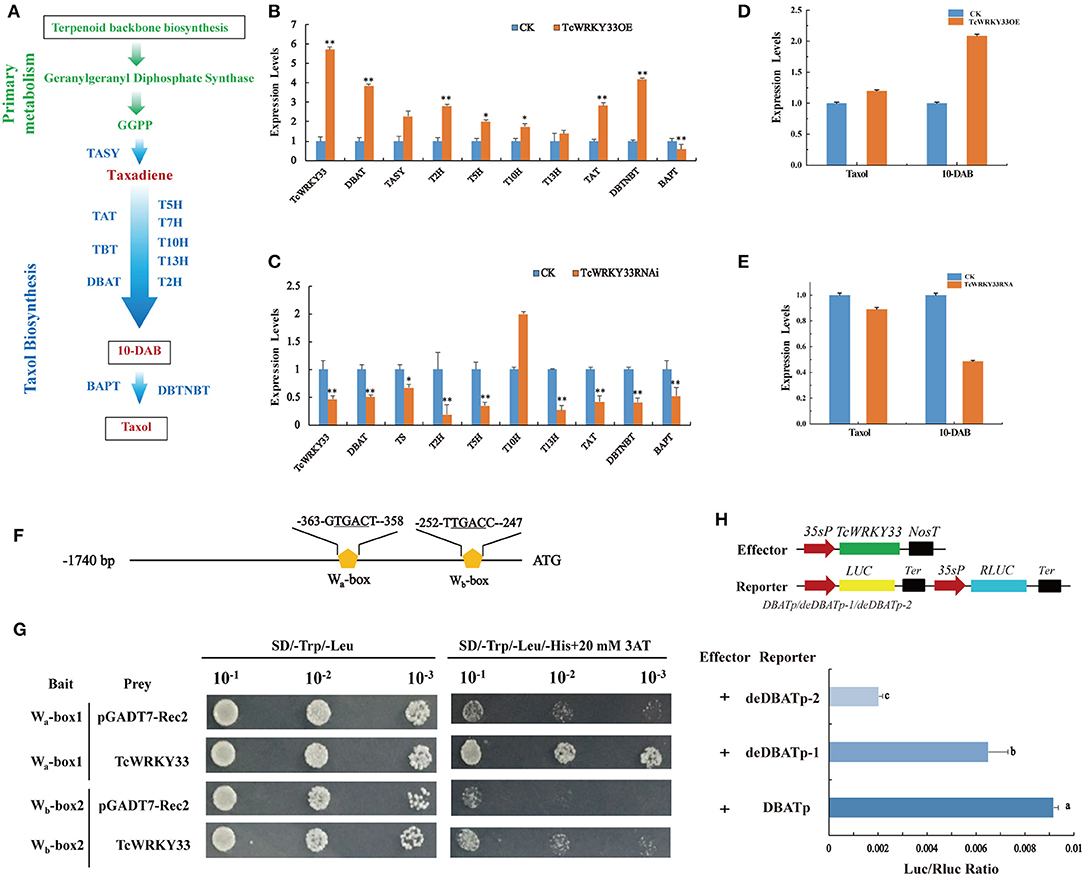
Figure 3. TcWRKY33 promoted the biosynthesis of taxol and 10-DAB by directly activating DBAT via binding with the W-boxes in its promoter. (A) A simplified diagram of taxol biosynthetic pathway. TASY, taxadiene synthase; T5H, taxadiene 5-alpha hydroxylase; T7H, 7-alpha-hydroxylase gene; T10H, 5-alpha-taxadienol-10-beta-hydroxylase; T13H, 13-alpha-hydroxylase gene; T2H, 2-alpha-hydroxylase gene; TAT, taxadienol acetyltransferase; TBT, taxane 2-alpha-O-benzoyltransferase; DBAT, 10-deacetylbaccatin III-10-O-acetyltransferase; BAPT, phenylpropanoyltransferase; DBTNBT, 3′-N-debenzoyltaxol N-benzoyltransferase. (B,C) The expression levels of taxol biosynthesis genes were quantified in TcWRKY33 overexpressed and RNA interference leaves by using qRT-PCR. Actin was used as the reference gene, each experiment was conducted three repeats, and totally three biological repeats were used, and the statistics method of p-value was Student's t-test. Asterisks indicate the significance, *0.01 < p < 0.05, and **p < 0.01. (D,E) The content of taxol and 10-DAB was quantified in TcWRKY33 overexpressed and RNA interference transient leaves by HPLC. The content of taxol in the CK group was as a reference. (F) Schematic diagram of DBAT promoter. The two orange pentagons indicated two W-boxes. (G) Yeast one hybrid of TcWRKY33 and W-boxes in yeast. Related bait vector with each W-box was co-transformed with vacant pGADT7-Rec2 into Y187 as the control. (H) Schematic diagrams of the effector vector and reporter vectors. TcWRKY33 expression plasmid was under the control of the CaMV 35S promoter. Effector and reporter vectors were co-transformed to tobacco leaves, and expression of LUC/Rluc was quantified. Different letters indicate the significant difference (p > 0.05) among treatments based on the Duncan's multiple range test of SPSS Statistics 21, SPSS Inc., Chicago, IL, USA.
We also interfered with TcWRKY33 expression in Taxus fresh leaves using transient RNAi technology. Expression levels of TcWRKY33 in RNAi leaves were 0.47 than that in the control (Figure 3C). Expression levels of taxol biosynthesis genes were measured by using qRT-PCR. Transcriptional levels of most of the taxol biosynthesis genes, including DBAT, TASY, T2H, T5H, 13-alpha-hydroxylase (T13H), TAT, and DBTNBT, were 0.50, 0.67, 0.19, 0.35, 0.37, 0.42, and 0.41 times those in the control; however, T10H displayed expression levels 2.00-fold those of the control (Figure 3C). These results confirmed that TcWRKY33 significantly upregulated the expression of taxol biosynthesis genes, especially the key biosynthesis genes DBAT and TASY. We selected these two genes for further study to explore the regulation mechanisms of TcWRKY33 on the taxol biosynthesis genes.
TcWRKY33 Stimulates the Accumulation of Taxol and 10-DAB
To further determine the effect of TcWRKY33 on taxol biosynthesis, we measured the content of taxol and 10-DAB (a taxol precursor and product of DBAT) using HPLC in T. chinensis leaves transiently overexpressing TcWRKY33 or with TcWRKY33 expression knocked down through RNA interference. Taxol accumulated to levels in TcWRKY33-overexpression leaves of up to 1.20 times higher than that in controls (Figure 3D). Furthermore, 10-DAB levels were up to 2.09 times higher than that in controls (Figure 3D). In TcWRKY33-RNA interference leaves, the content of taxol was only 0.89 than that in controls (Figure 3E). The accumulation of 10-DAB was also lower than that in controls by 0.49 (Figure 3E). These results indicate that the TcWRKY33 transcription factor effectively improved the biosynthesis of taxol. Moreover, TcWRKY33 seemed to be closely associated with the content of 10-DAB, which is a precursor of taxol produced by DBAT.
TcWRKY33 Directly Activates DBAT by Binding to W-boxes in Its Promoter
Li et al. (2013) discovered two W-boxes (Wa-box at −363 bp and Wb-box at −252 bp) within −368 bp of the DBAT promoter; these W-boxes are the functional sites of TcWRKY1, a group IIa WRKY factor (Li et al., 2013; Zhang et al., 2018a). DBAT, commonly considered one of the key enzymes in taxol biosynthesis, was significantly upregulated by TcWRKY33. Thus, DBAT appears to be a direct target of TcWRKY33. We analyzed the promoter of the DBAT gene for potential cis-elements regulated by TcWRKY33 (Figure 3F). Four tandem repeats of the two W-boxes (6 bp) were separately ligated into pHIS2.1 and respectively co-transformed with a TcWRKY33-AD fusion vector into yeast Y187 cells. Screening on SD/-Trp/-Leu/-His+20 mM 3-AT medium plates revealed that TcWRKY33 could bind to the two fragments (Figure 3G).
We further cloned the full-length 383 bp DBAT promotor and two progressive deletions removing the Wa-box and Wb-box into luciferase reporter vectors and, respectively, co-transformed these with a TcWRKY33 effector vector into N. benthamiana leaves (Figure 3H). LUC/Rluc values were decreased when W-boxes were deleted from the promotors. These results suggest that TcWRKY33 activates the expression of downstream genes by binding to the two W-boxes in the promoter of DBAT (Figure 3H). The LUC/Rluc ratio decreased more when both W-boxes were deleted than when only the Wa-box was deleted, indicating that the Wb-box might be the most important binding site for TcWRKY33 with the best activation activity (Figure 3H).
TcWRKY33 Activates the Activator of TASY by Binding to W-boxes in the TcERF15 Promoter
The expression of TASY, encoding one of the most important enzymes involved in taxol biosynthesis, was also activated by the TcWRKY33 transcription factor. However, this gene does not appear to possess a W-box in its promoter. This suggested the presence of an intermediate regulatory factor between TcWRKY33 and TASY. A search of reported regulators identified TcERF15, a positive regulator of the TASY gene that contains two W-boxes in its promoter (Zhang et al., 2015). After the induction of SA, the expression of TcERF15 upregulated by 2.66 times at 3 h (Figure 1B), indicating that TcERF15 responded positively to SA signals. When we transiently overexpressed TcWRKY33 in T. chinensis leaves, TcERF15 expression was slightly but significantly higher by 1.24 times compared with that in controls (Figure 4A). However, TcERF15 expression was on average 0.47 times that of controls in leaves transiently expressing a TcWRKY33 RNA interference construct, indicating that TcERF15 is probably involved in the regulation of TcWRKY33 (Figure 4A).
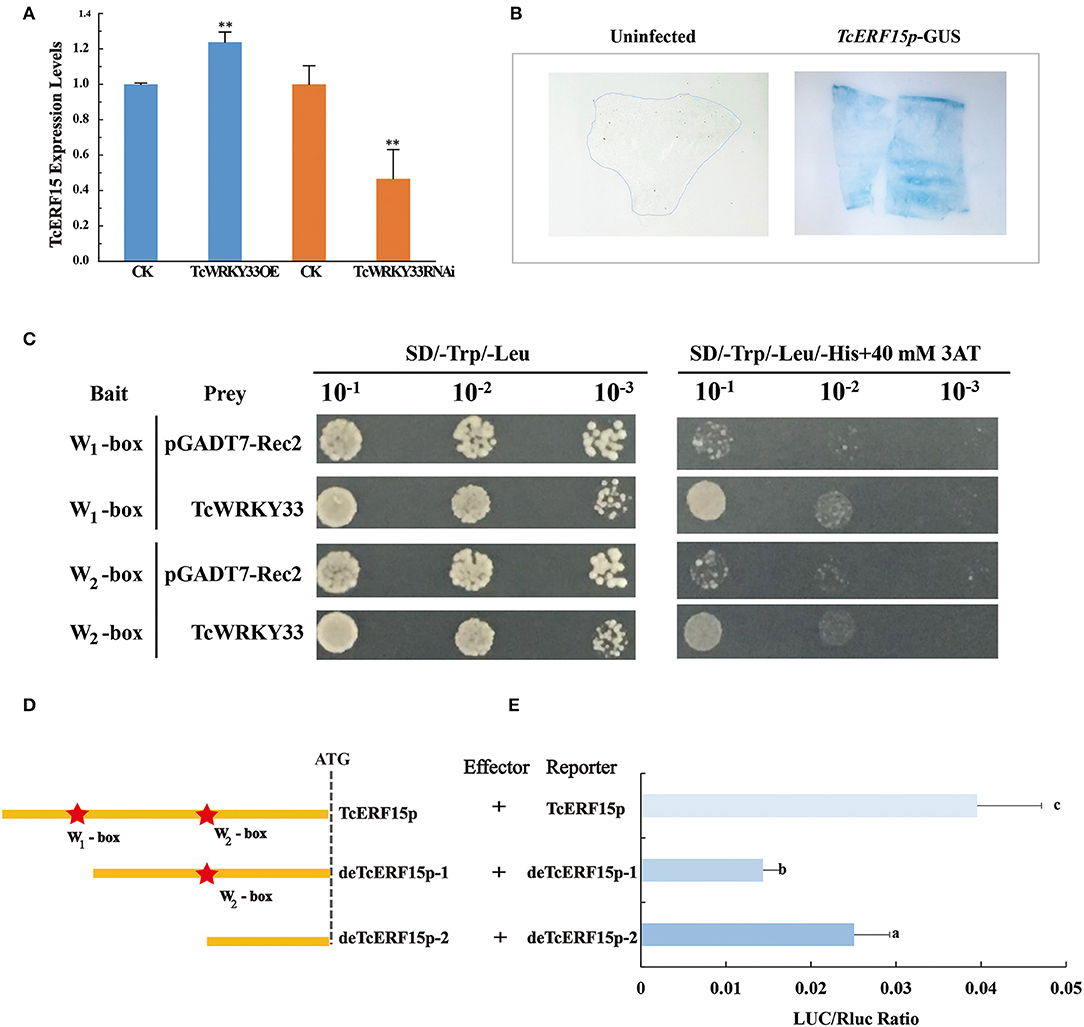
Figure 4. TcWRKY33 upregulated TASY by binding with the promoter of TcERF15. (A) The expression levels of TcERF15 were quantified in TcWRKY33 overexpressed and silenced leaves by using qRT-PCR. Asterisks indicate the significance, **p < 0.01. (B) GUS expression in transformed onion epidermis cells after 48 h. The untransformed onion epidermis cells as the negative control, the transformed onion epidermis cells with pBI121-proTcERF15::GUS was dyed blue by X-gluc. (C) Yeast one-hybrid analysis of interactions between TcWRKY33 and W-box. Yeast strains were serially diluted and 2.5 ml of each dilution was plated on synthetic dropout media without Leu and Trp (SD-L-W) plate or synthetic dropout media without Leu, Trp, and His (SD-L-W-H) plus 40 mM of 3-aminotriazole (3AT). (D) Schematic diagram of TcERF15 promoter. TcERF15 promoter contained two W-boxes that were signed by pentacles. This was the sketch map that the construction of full-length TcERF15 promotor and two progressive deletions removing the W1-box and W2-box. (E) Effector and reporter vectors were co-transformed, and the expression of LUC/Rluc was detected. Different letters indicate the significant difference (p > 0.05) among treatments based on the Duncan's multiple range test of SPSS Statistics 21.
We obtained a 715 bp 5′-flanking sequence of TcERF15. GUS staining indicated that this sequence could promote the expression of reporter genes and was thus a promoter fragment (Figure 4B). Online tools identified two W-boxes in the TcERF15 promoter: 5′-TTGACT-3′ at −700 bp (named W1-box) and 5′-TTGACG-3′ at −452 bp (named W2-box). We quadrupled each 6-bp fragment from the TcERF15 promoter and mutant fragments and ligated these separately into pHIS2.1; these constructs were, respectively, co-transformed with a TcWRKY33-AD fusion vector to detect interaction in a yeast one-hybrid system. Positive clones containing the TcWRKY33-AD fusion vector and W-box bait vectors grew on Trp-, Leu-, and His-deficient media with 40 mM 3-AT, whereas control clones did not. This indicated that TcWRKY33 interacts with W-boxes in the promoter of the TcERF15 gene (Figure 4C).
The yeast one-hybrid experiment revealed that the two W-boxes of the TcERF15 promoter were direct targets of TcWRKY33 in vitro. For further clarification, we transiently co-transformed full-length of TcERF15 promoter reporter vectors and two progressive deletion vectors, respectively, along with TcWRKY33 into N. benthamiana leaves (Figure 4D). LUC/Rluc expression was lower when the W1-box was deleted compared with expression from the full-length TcERF15 promoter, indicating that TcWRKY33 can activate the expression of reporter genes by binding to the W1-box. However, when both W-boxes were deleted, the LUC/Rluc ratio was higher than when only the W1-box was missing. This suggested that TcWRKY33 plays a negative regulatory role when interacting with the W2-box. The LUC/Rluc expression level was lower than that with the full-length promoter sequence, thus indicating that the activating function of the W1-box on TcWRKY33 is stronger than the negative regulatory role of the W2-box (Figure 4E).
Discussion
Taxol is a precious secondary metabolite that was initially isolated from Taxus spp. and is widely used as an anticancer drug (Rowinsky et al., 1990). However, it is found in extremely low levels in Taxus trees, making it too expensive to obtain in this manner (Walker and Croteau, 2000). A previous study using TcWRKY overexpression cell lines showed that these transcription factors may have significant effects on the regulation of taxol biosynthesis (Zhang et al., 2018a). SA also improves the accumulation of taxol (Miao et al., 2000). In this study, we isolated an SA-induced WRKY transcription factor, TcWRKY33, to explore its function in the biosynthesis of taxol.
Two W-boxes (Wa-box and Wb-box) in the promoter of DBAT are the main SA-responsive elements (Li et al., 2013). Thus, WRKY transcription factors must be key for transducing SA signals to the DBAT gene and possibly the whole taxol biosynthesis pathway. Despite interacting with the two W-boxes, however, TcWRKY1 only responds to MeJA signals (Li et al., 2013). We showed that TcWRKY33 responds strongly to SA, providing the missing SA-responsive WRKY regulator in taxol biosynthesis. Of seven TcWRKY transcription factors selected from transcriptome data to confirm their function in relation to SA and MeJA signals (Zhang et al., 2018a), only two were upregulated after SA treatment. They were also activated by MJ, which is considered to inhibit the SA response. TcWRKY33, therefore, presents a significant advantage as it is only induced by SA signals.
TcWRKY33 strongly prompted the accumulation of taxol and 10-DAB. Moreover, the expression levels of most taxol biosynthesis genes were increased when TcWRKY33 was overexpressed. This was particularly true for DBAT and TASY, which are recognized as the most important enzymes in taxol biosynthesis. The expression levels of these taxol biosynthesis genes were greatly decreased when TcWRKY33 was silenced. This revealed that TcWRKY33 functions as an active regulator of taxol biosynthesis by upregulating the expression of most taxol biosynthesis genes, including DBAT and TASY; this is consistent with other group I WRKY transcription factors functioning as activators (Zheng et al., 2006; Li et al., 2015).
Our yeast one-hybrid and dual-luciferase reporter experiments verified that TcWRKY33 activates the expression of DBAT by interacting with the two W-boxes in its promoter. The Wb-box showed greater activation activity than the Wa-box. Furthermore, the levels of the taxol precursor 10-DAB, a product of DBAT, also increased by up to 2.09 times after DBAT was activated by TcWRKY33. This confirms that TcWRKY33 increases the content of 10-DAB and taxol by directly activating DBAT expression. Similar results have been reported in other plants. For example, in Hylocereus monacanthus, HmoWRKY40 transcriptionally activates HmoCYP76AD by binding to its promoter and is involved in the regulation of pitaya betalain biosynthesis (Zhang et al., 2021). In Ophiorrhiza pumila, OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin by binding and activating the central camptothecin pathway gene OpTDC (Hao et al., 2021). WRKY transcription factors therefore influence the biosynthesis of secondary metabolites by regulating the expression of key enzyme genes in their biosynthesis pathways. However, TASY, one of the most important enzymes in the biosynthesis of taxol, which responds strongly to SA, appears to have no W-box in its promoter. Thus, TcWRKY33 upregulates its expression levels in other ways.
TcERF15, an activator of TASY, was activated in leaves transiently overexpressing TcWRKY33. Promoter analysis identified two W-boxes in the promoter of TcERF15. Yeast one-hybrid and dual-luciferase reporter assays further confirmed that TcWRKY33 activates TcERF15 by binding to the two W-boxes. Therefore, TcWRKY33 directly interacts with the W-boxes in the DBAT promoter to activate the expression of DBAT. It also increases the expression of TcERF15 slightly to upregulate TASY. Similar results have been observed in Catharanthus roseus. For example, CrWRKY1 plays a key role in determining the root-specific accumulation of serpentine by upregulating several key TIA pathway genes, especially tryptophan decarboxylase, as well as the transcriptional repressors ZCT1 (zinc-finger C. roseus transcription factor 1), ZCT2, and ZCT3 (Suttipanta et al., 2011). In red-fleshed apples, MdWRKY11 binds to W-boxes in the promoters of MdHY5, MdMYB10, and MdMYB11, regulating their activity. It also binds to the promoter of UFGT, encoding a key structural gene in the anthocyanin biosynthesis pathway, to upregulate the expression of UFGT. Hence, MdWRKY11 is involved in MdHY5-mediated anthocyanin biosynthesis and regulates the expression of MdMYB10, MdMYB11 transcription factors, and UFGT in apple fruit (Liu et al., 2019). These WRKY transcription factors obviously regulate the biosynthesis of secondary metabolites efficiently and extensively through various regulation patterns. Furthermore, our results show the superiority of TcWRKY33 in taxol biosynthesis.
TcWRKY33 clearly responds to SA signals and transmits these SA signals to downstream genes. In addition, TcWRKY33 appears to regulate downstream genes, such as DBAT and TASY, in two ways. DBAT responds to SA signals, which are transmitted directly to DBAT via TcWRKY33 interacting with W-boxes. The TASY gene is more sensitive to MeJA stimuli, and this mechanism has been clarified extensively (Lin et al., 1996). Additionally, TASY expression can be induced by SA, although the related mechanism remains unclear. In this study, we showed that TcWRKY33 exerts considerable positive effects on TASY expression, suggesting that TcWRKY33 may play important roles in the response of TASY to SA. As an activator of TASY, TcERF15 is a direct target of TcWRKY33. TcWRKY33 activates TASY by first regulating TcERF15 after the induction of SA. Thus, TcWRKY33 transmits SA signals to DBAT and TASY to regulate taxol biosynthesis (Figure 5).
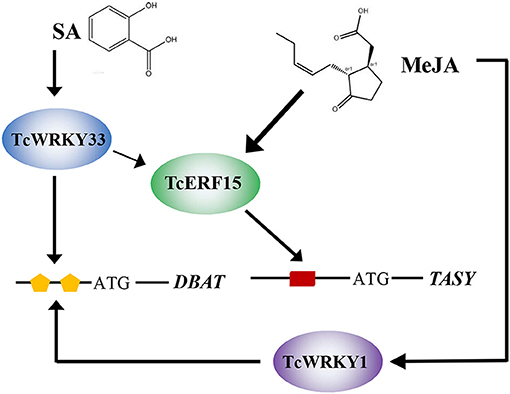
Figure 5. Model summarizing the TcWRKY33-mediated regulation of taxol biosynthesis in T. chinensis. The lines indicated the regulation. SA and MeJA, plant endogenous hormones; TcWRKY33 and TcWRKY1, T. chinensis WRKY transcription factors; TcERF15, T. chinensis ethylene responsive factor; orange pentagon and red rectangle represent W-boxes and GCC-box, cis-element in the gene promoter region, respectively; DBAT and TASY, taxadiene synthases.
Conclusion
We identified an SA-induced activator in the biosynthesis of taxol and provide references for the role of WRKY transcription factors in the regulation of taxol biosynthesis.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: NCBI repository, accession number: MW928850.
Author Contributions
CF, CZ, and LY conceived the research. YC and CZ designed the study. YC, HZ, MZ, WZ, ZO, and ZP conducted the experiments, including overexpression and RNA interference of TcWRKY33, taxanes content detection, subcellular localization, yeast one-hybrid, dual-luciferase reporter experiment, and so on. YC and HZ analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by the Major Technology Innovation of Hubei Province (2019ABA091). The funding bodies did not play a role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript, but just provide the financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the support in molecular biology experiments in Core Facilities of Life Sciences in the College of Science and Technology, Huazhong University of Science and Technology. We also thank the Analytical and Testing Center in the Huazhong University of Science & Technology for HPLC analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.697476/full#supplementary-material
Supplementary Table 1. Primers for amplifying genes, constructing vectors, and quantitative real-time PCR.
References
Abdul Rahim, R., Ahmad, N. H., Al Azzam, K. M., and Mat, I. (2018). Determination and quantification of the vinblastine content in purple, red, and white catharanthus roseus leaves using rp-hplc method. Adv. Pharm. Bull. 8, 157–161. doi: 10.15171/apb.2018.019
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on dna-binding selectivity of wrky transcription factors lend structural clues into wrky-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Croteau, R., Ketchum, R. E. B., Long, R. M., Kaspera, R., and Wildung, M. R. (2006). Taxol biosynthesis and molecular genetics. Phytochem. Rev. 5, 75–97. doi: 10.1007/s11101-005-3748-2
Cui, Y., Mao, R., Chen, J., and Guo, Z. (2019). Regulation mechanism of myc family transcription factors in jasmonic acid signalling pathway on taxol biosynthesis. Int. J. Mol. Sci. 20:1843. doi: 10.3390/ijms20081843
Duan, Y., Jiang, Y., Ye, S., Karim, A., Ling, Z., He, Y., et al. (2015). Ptrwrky73, a salicylic acid-inducible poplar wrky transcription factor, is involved in disease resistance in arabidopsis thaliana. Plant Cell Rep. 34, 831–841. doi: 10.1007/s00299-015-1745-5
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The wrky superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Hao, X., Xie, C., Ruan, Q., Zhang, X., Wu, C., Han, B., et al. (2021). The transcription factor opwrky2 positively regulates the biosynthesis of the anticancer drug camptothecin in ophiorrhiza pumila. Horticult. Res. 8:7. doi: 10.1038/s41438-020-00437-3
Kang, S., Jung, H., Kang, Y., Yun, D., Bahk, J., Yang, Y. K., et al. (2004). Effects of methyl jasmonate and salicylic acid on the production of tropane alkaloids and the expression of pmt and h6h in adventitious root cultures of scopolia parviflora. Plant Sci. 166, 745–751. doi: 10.1016/j.plantsci.2003.11.022
Kwak, S., Choi, M., Park, Y., Yoo, J., and Liu, J. (1995). Taxol content in the seeds of taxus spp. Phytochemistry 40, 29–32. doi: 10.1016/0031-9422(95)00247-5
Lan, A., Huang, J., Zhao, W., Peng, Y., Chen, Z., and Kang, D. (2013). A salicylic acid-induced rice (oryza sativa l.) transcription factor oswrky77 is involved in disease resistance of arabidopsis thaliana. Plant Biol. 15, 452–461. doi: 10.1111/j.1438-8677.2012.00664.x
Lenka, S. K., Nims, N. E., Vongpaseuth, K., Boshar, R. A., Roberts, S. C., and Walker, E. L. (2015). Jasmonate-responsive expression of paclitaxel biosynthesis genes in taxus cuspidata cultured cells is negatively regulated by the bhlh transcription factors tcjamyc1, tcjamyc2, and tcjamyc4. Front. Plant Sci. 6, 115. doi: 10.3389/fpls.2015.00115
Li, J., Wang, J., Wang, N., Guo, X., and Gao, Z. (2015). Ghwrky44, a wrky transcription factor of cotton, mediates defense responses to pathogen infection in transgenic nicotiana benthamiana. Plant Cell Tissue Organ Cult. 121, 127–140. doi: 10.1007/s11240-014-0688-9
Li, S., Zhang, P., Zhang, M., Fu, C., and Yu, L. (2013). Functional analysis of a wrky transcription factor involved in transcriptional activation of the dbat gene in taxus chinensis. Plant Biol. 15, 19–26. doi: 10.1111/j.1438-8677.2012.00611.x
Lin, X., Hezari, M., Koepp, A. E., Floss, H. G., and Croteau, R. (1996). Mechanism of taxadiene synthase, a diterpene cyclase that catalyzes the first step of taxol biosynthesis in pacific yew. Biochemistry 35, 2968–2977. doi: 10.1021/bi9526239
Liu, W., Wang, Y., Yu, L., Jiang, H., Guo, Z., Xu, H., et al. (2019). Mdwrky11 participates in anthocyanin accumulation in red-fleshed apples by affecting myb transcription factors and the photoresponse factor mdhy5. J. Agric. Food Chem. 67, 8783–8793. doi: 10.1021/acs.jafc.9b02920
Miao, Z. Q., Wei, Z. J., and Yuan, Y. J. (2000). Study on the effects of salicylic acid on taxol biosynthesis. Shengwu Gongcheng Xuebao 16, 509–513.
Nagata, T., Hara, H., Saitou, K., Kobashi, A., Kojima, K., Yuasa, T., et al. (2012). Activation of adp-glucose pyrophosphorylase gene promoters by a wrky transcription factor, atwrky20, in arabidopsis thaliana l. And sweet potato (ipomoea batatas lam.). Plant Product. Sci. 15, 10–18. doi: 10.1626/pps.15.10
Plevin, M. J., Mills, M. M., and Ikura, M. (2005). The lxxll motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30, 66–69. doi: 10.1016/j.tibs.2004.12.001
Robatzek, S., and Somssich, I. E. (2001). A new member of the arabidopsis wrky transcription factor family, atwrky6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. doi: 10.1046/j.1365-313X.2001.01131.x
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new endscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Rowinsky, E. K., Cazenave, L. A., and Donehower, R. C. (1990). Taxol: a novel investigational antimicrotubule agent. J. Natl. Cancer Inst. 82, 1247–1259. doi: 10.1093/jnci/82.15.1247
Schiff, P. B., Fant, J., and Horwitz, S. B. (1979). Promotion of microtubule assembly in vitro by taxol. Nature 277, 665–667. doi: 10.1038/277665a0
Sun, C., Palmqvist, S., Olsson, H., Boren, M., Ahlandsberg, S., and Jansson, C. (2003). A novel wrky transcription factor, susiba2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092. doi: 10.1105/tpc.014597
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor crwrky1 positively regulates the terpenoid indole alkaloid biosynthesis in catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tounekti, T., Hernández, I., and Munné-Bosch, S. (2013). Salicylic Acid Biosynthesis and Role in Modulating Terpenoid and Flavonoid Metabolism in Plant Responses to Abiotic Stress. Dordrecht: Springer, 141–162. doi: 10.1007/978-94-007-6428-6_8
Walker, K., and Croteau, R. (2000). Molecular cloning of a 10-deacetylbaccatin iii-10-o-acetyl transferase cdna from taxus and functional expression in escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 583–587. doi: 10.1073/pnas.97.2.583
Wang, F., Hou, X., Tang, J., Wang, Z., Wang, S., Jiang, F., et al. (2012). A novel cold-inducible gene from pak-choi (Brassica campestris ssp. Chinensis), bcwrky46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco. Mol. Biol. Rep. 39, 4553–4564. doi: 10.1007/s11033-011-1245-9
Wang, X., Guo, R., Tu, M., Wang, D., Guo, C., Wan, R., et al. (2017). Ectopic expression of the wild grape wrky transcription factor vqwrky52 in arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen botrytis cinerea. Front. Plant Sci. 8:97. doi: 10.3389/fpls.2017.00097
Wang, Y., Wu, J., and Yuan, Y. (2007). Salicylic acid-induced taxol production and isopentenyl pyrophosphate biosynthesis in suspension cultures of taxus chinensis var. Mairei. Cell Biol. Int. 31, 1179–1183. doi: 10.1016/j.cellbi.2007.03.038
Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P., and McPhail, A. T. (1971). Plant antitumor agents. Vi. The isolation and structure of taxol, a novel antileukemic and antitumor agent from taxus brevifolia. J. Am. Chem. Soc. 93, 2325–2327. doi: 10.1021/ja00738a045
White, N. J. (2008). Qinghaosu (artemisinin): the price of success. Science 320, 330–334. doi: 10.1126/science.1155165
Yukimune, Y., Tabata, H., Higashi, Y., and Hara, Y. (1996). Methyl jasmonate-induced overproduction of paclitaxel and baccatin III in taxus cell suspension cultures. Nat. Biotechnol. 14, 1129–1132. doi: 10.1038/nbt0996-1129
Zhang, L., Chen, C., Xie, F., Hua, Q., Zhang, Z., Zhang, R., et al. (2021). A novel wrky transcription factor hmowrky40 associated with betalain biosynthesis in pitaya (hylocereus monacanthus) through regulating hmocyp76ad1. Int. J. Mol. Sci. 22:2171. doi: 10.3390/ijms22042171
Zhang, M., Chen, Y., Nie, L., Jin, X., Liao, W., Zhao, S., et al. (2018a). Transcriptome-wide identification and screening of wrky factors involved in the regulation of taxol biosynthesis in taxus chinensis. Sci. Rep. 8:5197. doi: 10.1038/s41598-018-23558-1
Zhang, M., Jin, X., Chen, Y., Wei, M., Liao, W., Zhao, S., et al. (2018b). Tcmyc2a, a basic helix-loop-helix transcription factor, transduces ja-signals and regulates taxol biosynthesis in taxus chinensis. Front. Plant Sci. 9:863. doi: 10.3389/fpls.2018.00863
Zhang, M., Li, S., Nie, L., Chen, Q., Xu, X., Yu, L., et al. (2015). Two jasmonate-responsive factors, tcerf12 and tcerf15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in taxus chinensis. Plant Mol. Biol. 89, 463–473. doi: 10.1007/s11103-015-0382-2
Zhang, S., Li, C., Wang, R., Chen, Y., Shu, S., Huang, R., et al. (2017). The arabidopsis mitochondrial protease ftsh4 is involved in leaf senescence via regulation of wrky-dependent salicylic acid accumulation and signaling. Plant Physiol. 173, 2294–2307. doi: 10.1104/pp.16.00008
Keywords: SA signal, WRKY transcription factor, taxol biosynthesis, TcERF15, Taxus chinensis 3
Citation: Chen Y, Zhang H, Zhang M, Zhang W, Ou Z, Peng Z, Fu C, Zhao C and Yu L (2021) Salicylic Acid-Responsive Factor TcWRKY33 Positively Regulates Taxol Biosynthesis in Taxus chinensis in Direct and Indirect Ways. Front. Plant Sci. 12:697476. doi: 10.3389/fpls.2021.697476
Received: 19 April 2021; Accepted: 08 July 2021;
Published: 09 August 2021.
Edited by:
Sameh S. M. Soliman, University of Sharjah, United Arab EmiratesReviewed by:
Barunava Patra, University of Kentucky, United StatesSitakanta Pattanaik, University of Kentucky, United States
Copyright © 2021 Chen, Zhang, Zhang, Zhang, Ou, Peng, Fu, Zhao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunfang Zhao, cfzhao@mail.hust.edu.cn
†These authors have contributed equally to this work and share first authorship
 Ying Chen1,2,3†
Ying Chen1,2,3† Meng Zhang
Meng Zhang Chunhua Fu
Chunhua Fu