Predicting Present and Future Suitable Climate Spaces (Potential Distributions) for an Armillaria Root Disease Pathogen (Armillaria solidipes) and Its Host, Douglas-fir (Pseudotsuga menziesii), Under Changing Climates
- 1USDA Forest Service, Pacific Northwest Research Station, Corvallis, OR, United States
- 2USDA Forest Service, Rocky Mountain Research Station, Moscow, ID, United States
- 3Department of Agricultural Biology, Colorado State University, Ft. Collins, CO, United States
- 4USDA Forest Service, Southern Region, Atlanta, GA, United States
Climate change and associated disturbances are expected to exacerbate forest root diseases because of altered distributions of existing and emerging forest pathogens and predisposition of trees due to climatic maladaptation and other disturbances. Predictions of suitable climate space (potential geographic distribution) for forest pathogens and host trees under contemporary and future climate scenarios will guide the selection of appropriate management practices by forest managers to minimize adverse impacts of forest disease within forest ecosystems. A native pathogen (Armillaria solidipes) that causes Armillaria root disease of conifers in North America is used to demonstrate bioclimatic models (maps) that predict suitable climate space for both pathogen and a primary host (Pseudotsuga menziesii, Douglas-fir) under contemporary and future climate scenarios. Armillaria root disease caused by A. solidipes is a primary cause of lost productivity and reduced carbon sequestration in coniferous forests of North America, and its impact is expected to increase under climate change due to tree maladaptation. Contemporary prediction models of suitable climate space were produced using Maximum Entropy algorithms that integrate climatic data with 382 georeferenced occurrence locations for DNA sequence-confirmed A. solidipes. A similar approach was used for visually identified P. menziesii from 11,826 georeferenced locations to predict its climatic requirements. From the contemporary models, data were extrapolated through future climate scenarios to forecast changes in geographic areas where native A. solidipes and P. menziesii will be climatically adapted. Armillaria root disease is expected to increase in geographic areas where predictions suggest A. solidipes is well adapted and P. menziesii is maladapted within its current range. By predicting areas at risk for Armillaria root disease, forest managers can deploy suitable strategies to reduce damage from the disease.
Introduction
Armillaria root disease, caused by Armillaria spp., is a leading cause of growth loss and mortality of a diverse range of horticultural and timber trees in many temperate regions around the world (e.g., Heinzelmann et al., 2019). In coniferous forests of North America, Armillaria solidipes (formerly North American A. ostoyae) is a native pathogen that causes Armillaria root disease that negatively impacts growth and survival of many conifer species (Ferguson et al., 2003; Lockman and Kearns, 2016) including Pseudotsuga menziesii (Douglas-fir), which is a dominant component of many forest stands in western North America and an important tree for ecological, sociological, and economic purposes.
Changing climate, extreme weather events (e.g., drought, high temperatures), and/or other disturbances (e.g., fire, insect attack, and forest management activities) are causing increased disease severity by A. solidipes on stressed P. menziesii (Morrison, 2011; Murray and Leslie, 2021). Armillaria root disease, in conjunction with the adverse impacts from climate and extreme weather events, can predispose trees to attack by other biotic agents including bark beetles (e.g., Hertert et al., 1975; Kulhavy et al., 1984; Tkacz and Schmitz, 1986), which can result in tree mortality and increased availability of nutritional substrates for inoculum of Armillaria species that can, in turn, contribute to future disease on the site. When characterizing the ecological role of Armillaria in western North America, it is crucial to consider that Armillaria also behaves as a saprophyte, colonizing trees that succumbed to other abiotic and biotic causes. Thus, careful observations are required to determine the interactive role of Armillaria root disease in relation to climate, extreme weather, and other interacting biotic/abiotic factors (Kubiak et al., 2017). While Armillaria root disease is well known for its negative impacts on tree health, causing economic impact and reduced carbon sequestration potential of forests, it is important to note that Armillaria can also contribute ecological benefits, which include nutrient cycling, habitat formation (e.g., nesting cavities), and food for wildlife, such as small birds and mammals (e.g., Steeger and Hitchcock, 1998; Parsons et al., 2003; Heinzelmann et al., 2019).
Although information is generally lacking on the incidence and spread of Armillaria root disease over time and space in association with climate and extreme weather events, the known spatial occurrence of A. solidipes from extensive sampling offers a unique opportunity to apply bioclimatic modeling to identify climatic components associated with its occurrence. Output from these models can be coupled with GIS to project maps that show the predicted suitable climate space (potential distribution or realized climate niche) across geographic areas, and when coupled with output from general circulation models (GCMs) show predicted future distributions of suitable climate spaces for P. menziesii and A. solidipes. Understanding the host-pathogen interaction under changing climates allows for predictions of the potential distributions of P. menziesii and A. solidipes across current and future landscapes to inform Armillaria root disease management.
Armillaria occurs primarily below ground, which represents a distinct environment that is relatively stable from minor weather fluctuations. Below-ground conditions (e.g., soil temperature and moisture) and microclimate can affect A. solidipes, soil microbial communities, and/or P. menziesii. These below-ground, climatic conditions are influenced by latitude/longitude, elevation, slope, aspect, topography, drainage, soil properties, crown cover, etc. The distribution of Armillaria root disease on a site is largely attributable to soil environments and microclimates that vary within and/or among stands. In addition, the occurrence of A. solidipes is associated with plant association groups or habitat types (i.e., well-defined associations of trees, shrubs, and herbaceous plants) that reflect soil temperature (e.g., represented by overstory trees) and moisture (e.g., represented understory plants) in conifer forests of western North America (e.g., McDonald et al., 2000; Kim et al., 2010). At the broadscale, bioclimatic models (e.g., MaxEnt: Maximum entropy species distribution modeling) can use presence-only survey data to project potential distribution of both pathogen and host tree species based on major climatic factors (e.g., 19 bioclimatic variables from the WorldClim) for contemporary or projected future time periods (Phillips et al., 2006). Furthermore, climate layers of projected future climates based on different greenhouse emission scenarios can be used to predict future potential distribution of both pathogen and host tree species under different projected climate scenarios.
In western North America, Armillaria root disease caused by A. solidipes is common on trees that are ecophysiologically maladapted (stressed by environmental factors), which can be attributed to climatic maladaptation and/or other disturbances (McDonald et al., 1987a; McDonald, 1990; Murray and Leslie, 2021). Projections of climate change in the northwestern United States predict an increase in the frequency and intensity of drought and higher temperatures that will result in more stressful environmental conditions for forest trees (Chmura et al., 2011). Thus, a reasonable approach toward predicting climate change impacts on Armillaria root disease at the broad scale is to use bioclimatic models to predict the realized climatic niche for the P. menziesii and A. solidipes pathosystem (e.g., Rehfeldt et al., 2006; Klopfenstein et al., 2009). Although P. menziesii spans a wide geographic (19–44°N latitude and 100–118°W longitude; Hermann and Lavender, 1990) and climatic range in western North America, it is considered a specialist in terms of its climatic requirements for populations, making individual stands especially vulnerable to climate-induced stresses (e.g., Weiskittel et al., 2012; Rehfeldt et al., 2014b).
Our goals were to (1) predict suitable climate spaces (potential distributions) for P. menziesii and A. solidipes across western North America for contemporary climate and projected climate for the average of years 2081–2100; and (2) draw inferences from comparisons of the prediction models for the host (P. menziesii) and pathogen (A. solidipes) to predict geographic areas where P. menziesii will become maladapted due to climate change, which will likely exacerbate Armillaria root disease in areas where A. solidipes remains climatically adapted. We used georeferenced occurrence locations for visually identified P. menziesii and DNA sequence-confirmed A. solidipes in combination with location-specific climate data for bioclimatic modeling to predict where A. solidipes is likely to occur and cause increased disease pressure on maladapted and/or stressed P. menziesii under changing climatic conditions.
Materials and Methods
Pseudotsuga menziesii Location Data
A total of 11,826 locations of P. menziesii were used from selected “fuzzy” coordinates obtained from Forest Inventory Analyses (FIA) data (Rehfeldt et al., 2014a) within the continental United States (locations from Canada and Mexico were not included because the FIA data are only available for the contiguous United States) (Figure 1A).
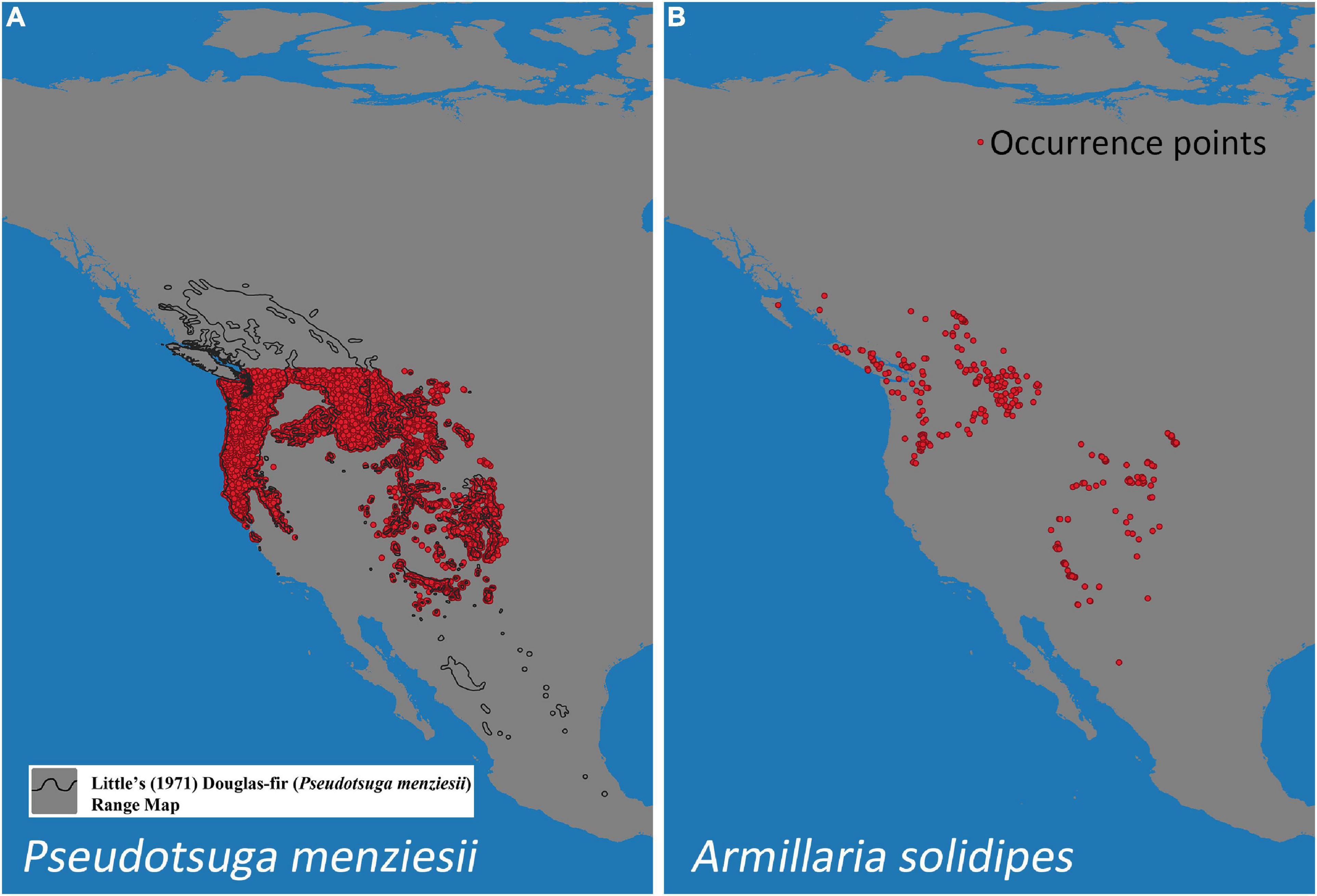
Figure 1. Occurrence points of Pseudotsuga menziesii (A) and Armillaria solidipes (B) in North America. The black outline (A) designates Little’s (1971) P. menziesii range map.
Armillaria solidipes Surveys, Identification, and Location Data
Briefly, surveys for Armillaria root disease pathogens require collection of Armillaria mycelia (e.g., mycelial fan, rhizomorph, or basidiocarp) for Armillaria identification via DNA sequences (e.g., Kim et al., 2000, 2006; Ross-Davis et al., 2012; Elías-Román et al., 2013; Klopfenstein et al., 2017). Careful examination of symptoms and signs can confirm that Armillaria is acting as a pathogen on the host. In addition, precise locations of identified Armillaria collections were recorded so that bioclimatic modeling can be applied to determine climatic influences on Armillaria root disease pathogens.
Armillaria solidipes point locations were collected from previous studies of distribution and ecology from the states/provinces of Washington, Oregon, Idaho, Montana, Utah, Wyoming, South Dakota, Arizona, Colorado, New Mexico, British Columbia (BC, Canada), and Chihuahua (Mexico) (McDonald et al., 1987b,2011; Shaw, 1989; Omdal et al., 1995; McDonald, 1998, unpublished data; Kim et al., 2000; Ferguson et al., 2003; Van der Star et al., 2003; Worrall et al., 2004; Hanna et al., 2007, 2008, 2009, 2014; Blodgett and Lundquist, 2011; Klopfenstein et al., 2012; Berbee et al., 2014, 2015; Hoffman et al., 2014; Blodgett et al., 2015 and Hanna, unpublished data). For Armillaria surveys, the main lateral roots of diverse trees were excavated and inspected, and Armillaria samples were collected as mycelial fans under the bark, rhizomorphs, basidiocarps, and/or decayed wood. Isolates were established in culture and were confirmed as A. solidipes using basidiocarp morphology (e.g., some Canadian locations) and DNA-based species identification (e.g., Kim et al., 2000, 2006; Ross-Davis et al., 2012; Elías-Román et al., 2013; Klopfenstein et al., 2017). From these studies, A. solidipes isolates were recorded from 382 distinct locations throughout western North America (Figure 1B).
Climate Data and Bioclimatic Variables
Climate-based, species-distribution models using MaxEnt and 19 bioclimatic variables (Table 1) from Worldclim version 2.1 (Fick and Hijmans, 2017) were created for both host (P. menziesii) (Figure 2) and pathogen (A. solidipes) (Figure 3) for the contemporary (1970–2000) and future time periods (2081–2100) using Coupled Model Intercomparison Project phase 6 (CMIP6) (Eyring et al., 2016) data consisting of two different Shared Socioeconomic Pathways (SSPs) paired with The Canadian Earth System model version 5 (CanESM5) GCM (Swart et al., 2019) using 2.5-min data. In general, five SSPs narratives describe the broad socioeconomic trends that could shape future society and we chose to use two scenarios: SSP2-4.5 (“middle of the road” world where trends do not shift markedly from historical patterns) and SSP5-8.5 (“fossil-fueled development,” which follows path of rapid and unconstrained growth in economic output and fossil fuel-based energy use). In addition, change in climate suitability for P. menziesii from the contemporary time period to the future time period for both SSPs (SSP2-4.5 and SSP5-8.5) were visually compared (Figure 4).
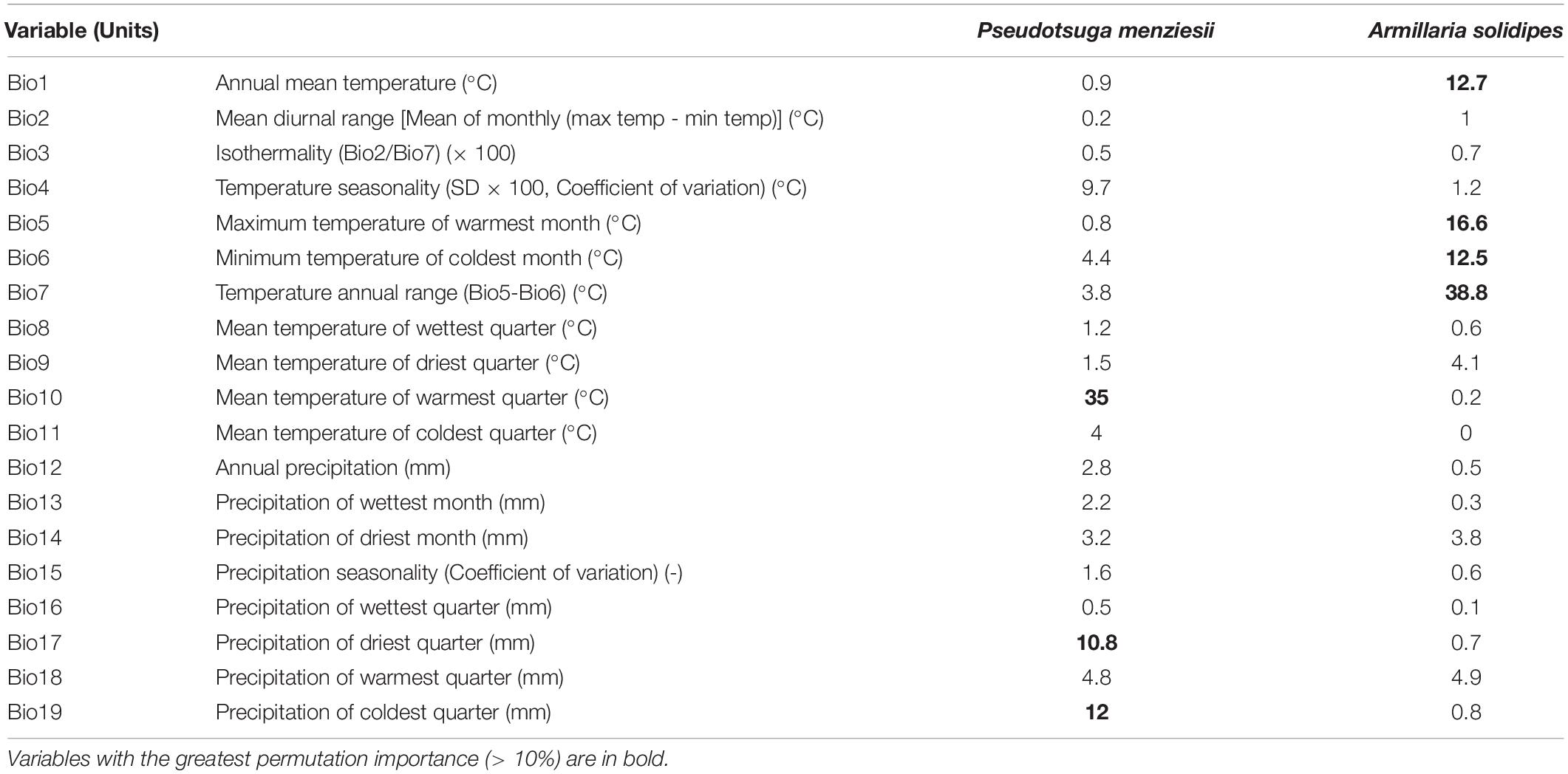
Table 1. Summary of MaxEnt’s permutation importance value for each of the contemporary 19 bioclimatic variables (Bio1–Bio19) derived from the WorldClim (worldclim.org) database (1970–2000) used to model suitable climate space (potential distribution) based on occurrence points of Pseudotsuga menziesii and Armillaria solidipes.
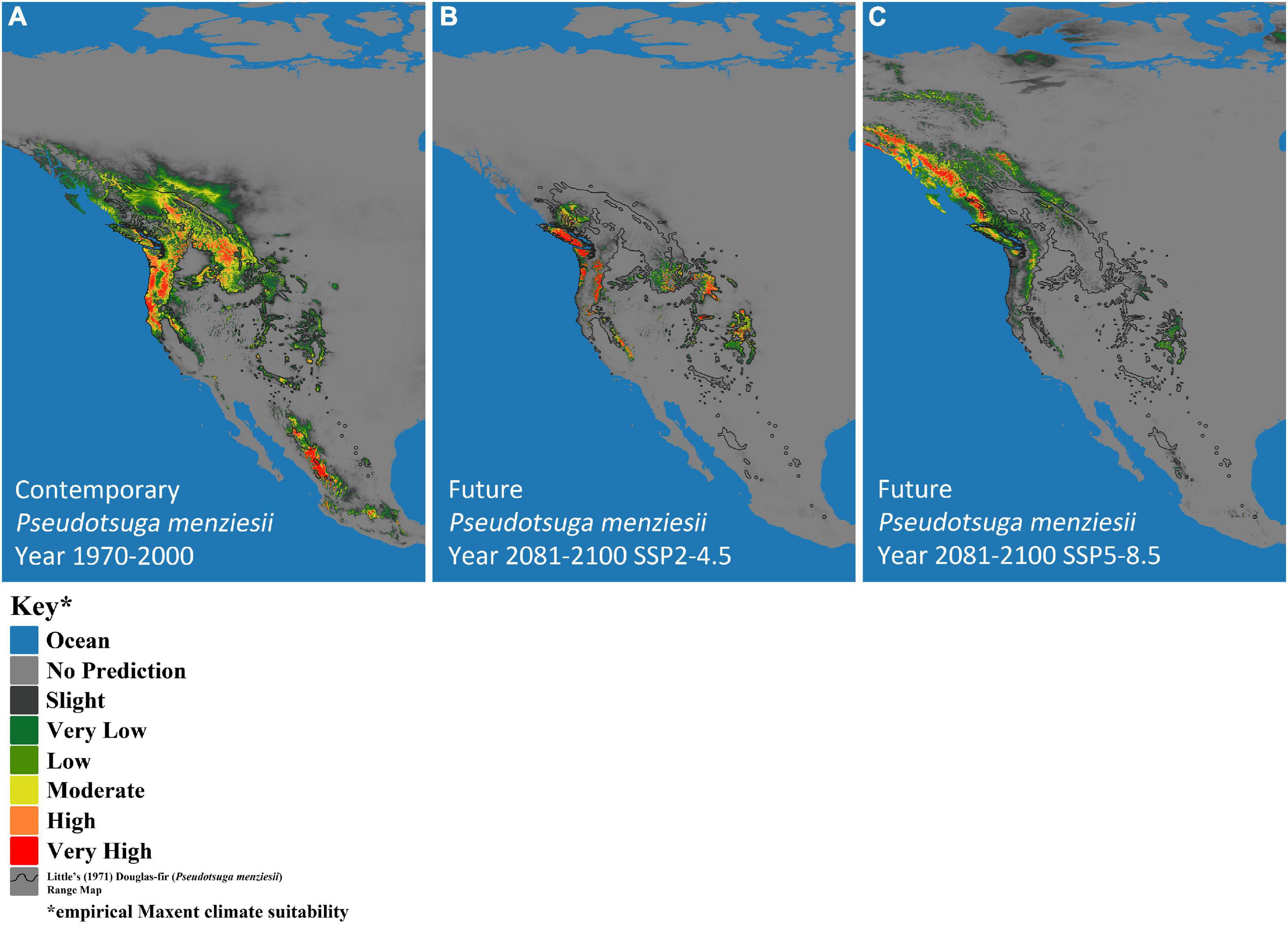
Figure 2. MaxEnt-based, bioclimatic models for Pseudotsuga menziesii (Douglas-fir) in western North America. (A) Contemporary years (1970–2000), and two future models show two Shared Socioeconomic Pathway (SSP) scenarios (SSP2-4.5 and SSP5-8.5), (B) SSP2-4.5 and (C) SSP5-8.5. Darkest gray represents predicted empirical MaxEnt climate suitability, with dark green, light green, yellow, orange, and red indicating increased probability of climatic suitability, respectively. The black outline designates Little’s (1971) P. menziesii range map.
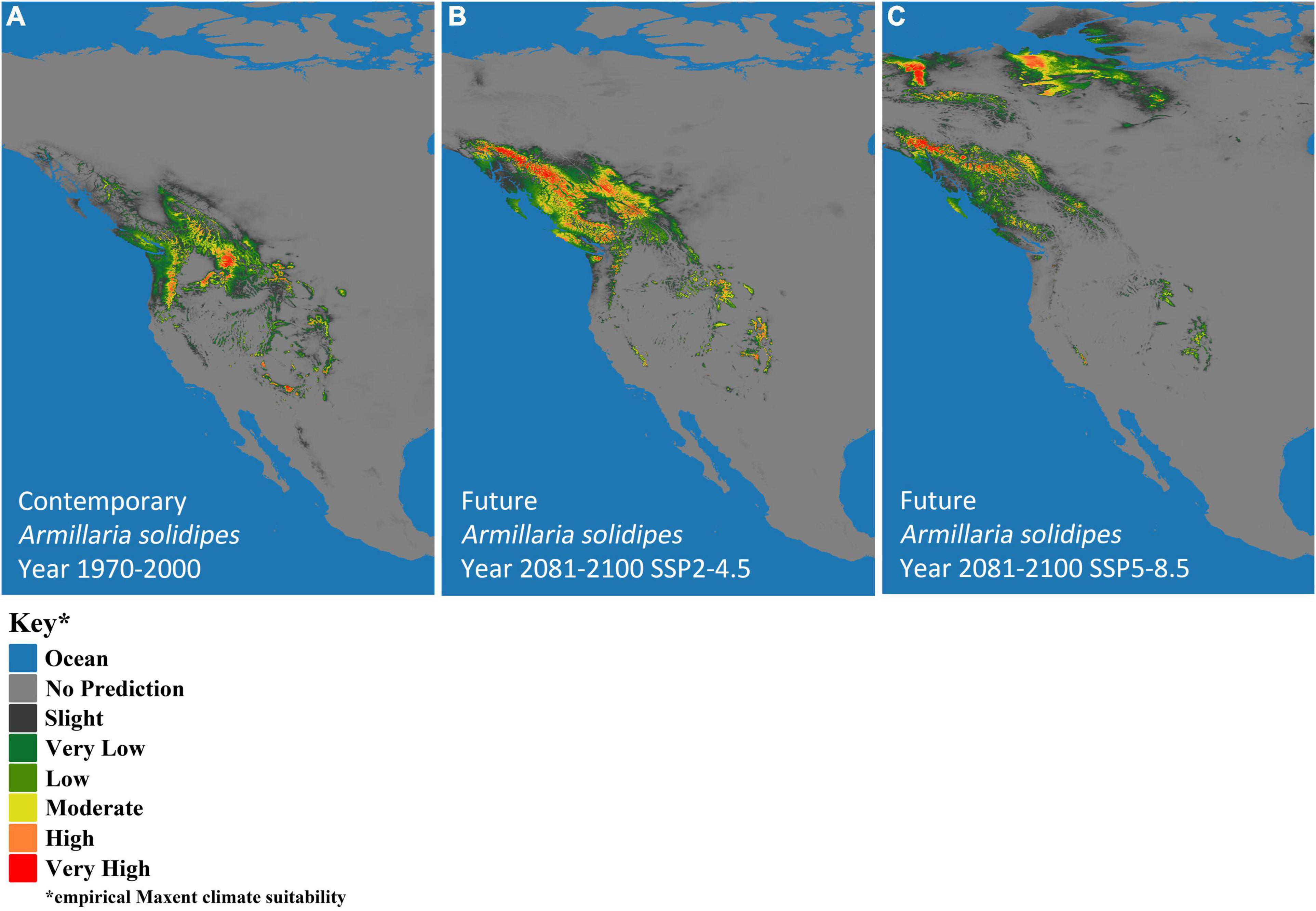
Figure 3. MaxEnt-based, bioclimatic models for Armillaria solidipes in western North America. (A) Contemporary years (1970–2000) and two future models show two Shared Socioeconomic Pathway (SSP) scenarios (SSP2-4.5 and SSP5-8.5), (B) SSP2-4.5 and (C) SSP5-8.5. Darkest gray represents predicted empirical MaxEnt climate suitability, with dark green, light green, yellow, orange, and red indicating increased probability of climatic suitability, respectively.
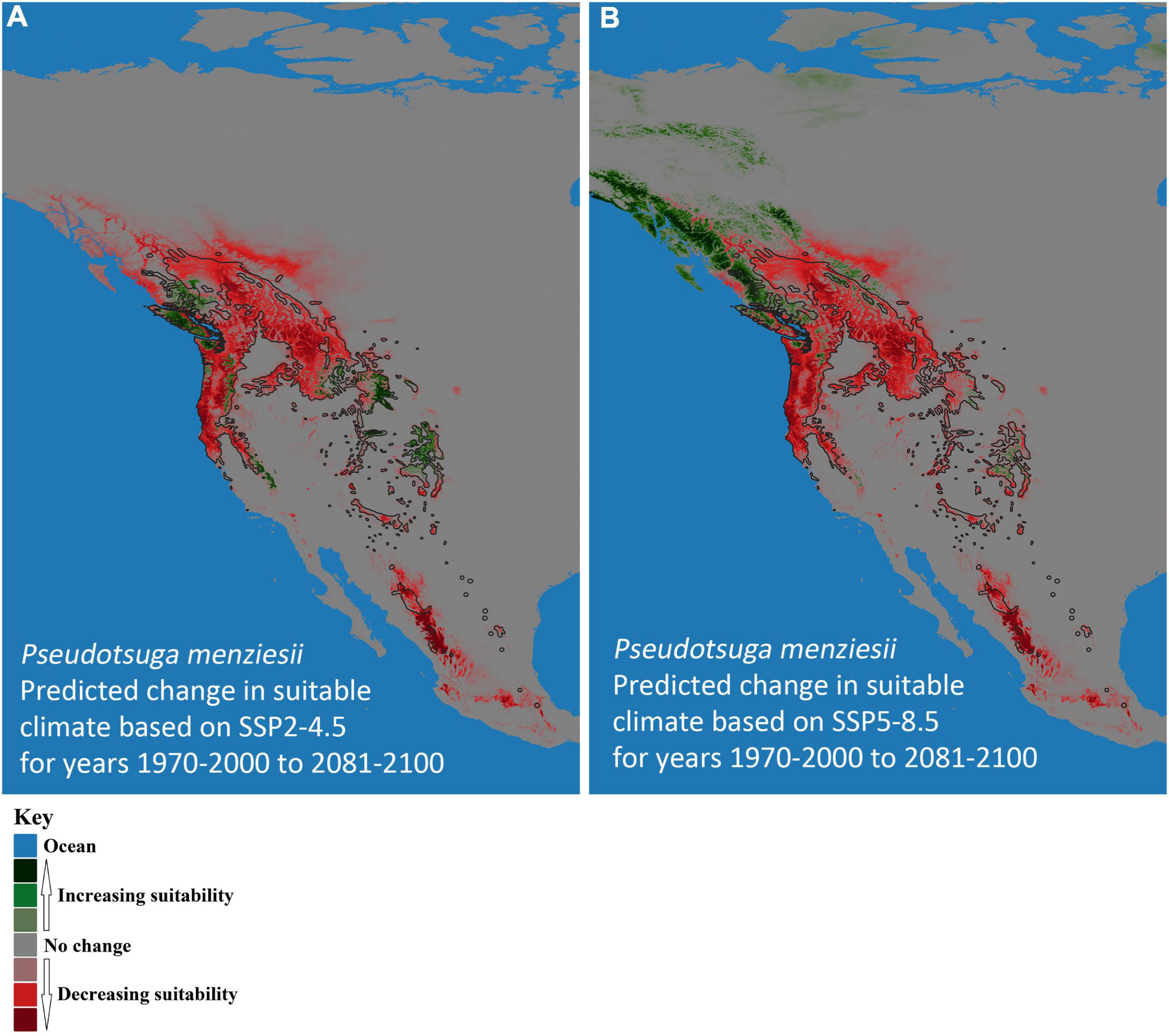
Figure 4. Predicted change in climate suitability for Pseudotsuga menziesii (Douglas-fir) in western North America from contemporary (1970–2000) to future (2081–2100) models. (A) Shared Socioeconomic Pathway (SSP) SSP2-4.5 and (B) SSP5-8.5. Pink to dark red, respectively, indicates increasing probability of disappearing climate suitability (i.e., ecophysiologically maladapted trees), while light green to dark green, respectively, represents predictions of new climatically suitable areas. The black outline designates Little’s (1971) P. menziesii range map.
MaxEnt Modeling
To allow calculations in suitability models, input data for MaxEnt consisted of SWD (samples with data) files for each species that linked geographic coordinates (presence point locations) with climate variable values for each of the 19 bioclimatic variables. The models used 19 bioclimatic variables in three sets of 2.5-min, interpolation grids from worldclim.org (Fick and Hijmans, 2017). MaxEnt also uses SWD files of background locations or “pseudo-absences” to “train” the models. For the P. menziesii model, 292,639 actual absence point locations were used as background data. MaxEnt’s logistic output (an index of probability from 0 to 1) was chosen for easier conceptualization compared to MaxEnt’s raw exponential model. For the A. solidipes model, background points were created from 10,000 randomly selected geographical locations within the geographic range of the collected isolates.
MaxEnt parameters included cross-validation with 20 replicate runs to generate statistical data. Quantum GIS (QGIS)1 was used to create the final outputs using MaxEnt’s cumulative output. Each cumulative value is the sum of probabilities of cells less than or equal to the cell grid, times 100. MaxEnt’s cumulative output was chosen for easier conceptualization compared to MaxEnt’s logistic (an index of probability from 0 to 1) or raw exponential output. Totals of 11,826 and 382 locations were used for P. menziesii and A. solidipes, respectively.
To evaluate the reliability of the model, the receiver operating characteristic (ROC) curve of the jackknife method was used (Dorfman et al., 1992). The areas under the ROC curve (AUC) values (ranging from 0 to 1) were generated to calculate model performance (e.g., prediction accuracy) based on true-positive, false-positive, true-negative, and false-negative rates. Higher AUC values (> 0.9) indicate higher reliability of the model (Wiley et al., 2003). Permutation importance (resulting from the values of each environmental variable on training presence and background data being randomly permuted, the model then re-evaluated on the permuted data; resulting drop in training AUC is normalized to percentages) of 19 bioclimatic variables derived from the WorldClim database used to model suitable climate space (potential distribution) based on known occurrence points for P. menziesii and A. solidipes (Table 1).
Results
The MaxEnt bioclimatic model using all occurrence records of P. menziesii (N = 11,826) and A. solidipes (N = 382) (Figures 1A,B, respectively) shows a prediction where many geographic areas throughout the North America have some degree of probability of suitable climate (i.e., possesses a climate suitable for species survival) for both P. menziesii and A. solidipes (Figures 2, 3, respectively). With 1.0 representing a perfect value and 0.5 representing presence by chance, the average test AUC ± standard deviation values, based on 20 replicate runs, were 0.953 ± 0.002 for P. menziesii and 0.960 ± 0.010 for A. solidipes.
For the P. menziesii model, the major bioclimatic variables (i.e., > 10%) of permutation importance were mean temperature of warmest quarter (Bio10), precipitation of coldest quarter (Bio19), and precipitation of driest quarter (Bio17). For the A. solidipes model, temperature annual range (Bio7), maximum temperature of warmest month (Bio5), annual mean temperature (Bio1), and minimum temperature of coldest month (Bio6) were the major bioclimatic variables (Table 1).
The predicted contemporary distribution of P. menziesii is consistent with the actual range of this tree species reported by Little (1971) (Figure 2A). The predicted contemporary distribution of A. solidipes shows a geographically wide distribution with more suitable habitat in the wetter forests of the Pacific Northwest and with pockets in the southwestern United States, which appear to be associated with areas that have monsoon seasons (Figure 3A). In general, the bioclimatic models predict that the contemporary suitable climatic space (potential distribution) of A. solidipes has considerable overlap with that of P. menziesii (e.g., northern Idaho, northern Cascade Mountains, Rocky Mountains, etc.).
The models predicted future suitable climate spaces for Year 2081–2100 under two climate scenarios, SSP2-4.5 and SSP5-8.5, are shown in Figures 2B,C, 3B,C respectively. For both P. menziesii and A. solidipes, the suitable habitat shifts northwest toward more northern and coastal areas. Notably, for P. menziesii, the suitable climate space shifts considerably northward and is much smaller in size under the SSP5-8.5 model compared to the SSP2-4.5 model (Figures 2B,C). Predicted changes in climate suitability for P. menziesii in western North America from contemporary (1970–2000) to future (2081–2100) models for both SSPs (SSP2-4.5 and SSP5-8.5) are shown in Figures 4A,B. Both future climate scenarios indicate increasing probability of disappearing climate suitability (i.e., ecophysiologically maladapted trees) for P. menziesii within its current range. Distributions of suitable climate space for P. menziesii in the Rocky Mountains that can be observed in Figure 4A are conspicuously absent under the more severe climate-change model of SSP5-8.5 (Figure 4B), though some climate suitability is observed in northern coastal regions.
When comparing the SSP2-4.5 and SSP5-8.5 models for A. solidipes, differences in suitable climate space are striking. In Figure 3B (SSP-4.5), the suitable climate space shifts northward and toward coastal areas, fairly similar to the P. menziesii suitable climate space predicted under the SSP2-4.5 model. However, the suitable climate space predicted under the SSP5-8.5 model highlights a new northern region of climatic suitability that coincides with newly identified areas with potential climatic suitability for P. menziesii under the same model (Figure 3C).
Discussion
This study provides a sound approach for predicting potential forest disease incidence and potential severity under contemporary and changing climate scenarios. We used presence-only occurrence data for A. solidipes and both presence and absence (as background data) occurrence data for P. menziesii to produce predictions of potential contemporary and future distributions of these species using MaxEnt modeling. Predicted shifts in suitable climate space for both P. menziesii and A. solidipes are highly correlated as the predicted change in climate suitability moves dramatically northward for years 2081–2100 using both SSPs (SSP2-4.5 and SSP5-8.5). The results from this study strongly suggest that Armillaria root disease will be exacerbated in many areas where P. menziesii is predicted to become increasingly maladapted and extreme weather events will likely become more common. Predicting the potential distribution of both pathogen (A. solidipes) and a primary host (P. menziesii) under contemporary and future climate scenarios is critical to deploy suitable strategies to reduce damage from the disease.
In this study, the MaxEnt models performed remarkably well, based on the high AUC values (0.953 for P. menziesii and 0.960 for A. solidipes) that indicate high reliability and predictive accuracy. This approach seems to be quite reliable, especially given the relatively limited occurrence data for A. solidipes. In addition, MaxEnt is a robust predictor of collinearity in model training and highly correlated variables have little impact on the model output (Feng et al., 2019). This study predicts areas at risk for Armillaria root disease using the MaxEnt model, which works well with limited number of presence-only occurrence data (Phillips et al., 2006). Unlike P. menziesii (N = 11,826), the acquisition of occurrence data for A. solidipes (N = 382) required highly labor-intensive work including excavation of lateral roots, pathogen isolation, and DNA sequence-based identification to confirm occurrence (Klopfenstein et al., 2009). In addition, absence data for Armillaria are difficult to obtain and confirm with certainty. For these reasons, MaxEnt was applied to compare both host (P. menziesii) and pathogen (A. solidipes) using the same modeling parameters, although the model slightly overpredicts the suitable climate space compared to a presence/absence model (see Rehfeldt et al., 2014a for a climate niche model of P. menziesii based on a Random Forests classification algorithm). Overall, under the contemporary climate, the predicted potential distribution of P. menziesii using the MaxEnt is generally congruent with the established range map of P. menziesii within the continental United States (Little, 1971). Recently, MaxEnt bioclimatic models have been widely used to investigate the suitable climate space of other plants and plant pathogens, with high predictive performance (e.g., Kumar and Stohlgren, 2009; Khanum et al., 2013; Remya et al., 2015; Stewart et al., 2018, 2020; Tang et al., 2021).
Several other critical variables can be implemented to improve the precision of these prediction models for P. menziesii and A. solidipes. For P. menziesii, the MaxEnt models were based on presence and absence data at the species level. However, P. menziesii exhibits climatic adaptation at the intraspecies level where a given population within the species is genetically adapted to persist only over a portion of the climate range occupied by the species as a whole (e.g., Weiskittel et al., 2012; Rehfeldt et al., 2014a). Because our bioclimatic models do not explicitly include intraspecies adaptive structure (i.e., climate types), they likely underestimate the extent of climatic maladaptation within the species under predicted climate change. Of special note, P. menziesii with higher levels of drought tolerance were also associated with higher levels of resistance to Armillaria root disease, caused by A. solidipes (Cruickshank and Filipescu, 2017). Furthermore, summer droughts were associated with increased A. solidipes-caused mortality of P. menziesii in southern BC, Canada (Murray and Leslie, 2021). These findings further support that climatic maladaptation of P. menziesii will lead to increased infection by A. solidipes. Thus, predictions of P. menziesii maladaptation under future climate scenarios likely underestimate the extent of maladaptation and associated increased susceptibility to Armillaria root disease. However, east of the Cascade Range, P. menziesii is one of the most susceptible tree species to Armillaria root disease (Morrison, 1981; Robinson and Morrison, 2001), but the faster growing coastal variety of P. menziesii succumbs less often to A. solidipes infection (Johnson et al., 1972; Robinson and Morrison, 2001). In addition, the population structure of A. solidipes is unknown; however, an individual A. solidipes genet (vegetative clone) can reside on a site up to ca. 1900–8650 years (Ferguson et al., 2003), suggesting that this root pathogen may possess a broad tolerance for changing climatic conditions. To address genetic differences within the species, population-level data are needed for both host and pathogen to refine climate-based models. Additional predictive variables (e.g., soil types, soil temperature and moisture, drainage properties/patterns, solar radiation, predictions of other Armillaria species, soil microbial communities, etc.) will affect the suitable climate distribution of A. solidipes, and this information could improve the resolution of predictive maps.
Reliable, long-term records are unavailable for quantifying Armillaria root disease levels because of difficulties associated with below-ground surveying, sampling, and identifying Armillaria spp. Long-term monitoring for Armillaria root disease in relation to weather/climate factors would provide an opportunity to directly determine climatic influences on the impact of the disease; however, such studies are complex because tree maturity and other stand conditions also change over time. Long-term monitoring is needed to further establish any climatic relationships with P. menziesii maladaptation, A. solidipes growth/survival, and Armillaria root disease of P. menziesii. Monitoring must also consider that the majority of P. menziesii associated with Armillaria root disease show no readily observable symptoms, except slower growth (Morrison et al., 2000; Cruickshank et al., 2011). In addition, evidence suggests complex interactions among Armillaria spp., in which some Armillaria sp. may serve as a biocontrol against more pathogenic Armillaria sp. under favorable site conditions (e.g., A. altimontana vs. A. solidipes) (Hanna, 2005; Warwell et al., 2019). For this reason, bioclimatic modeling of Armillaria spp. that are currently existing as saprophytes in a non-pathogenic mode is also warranted. Monitoring data need to include presence/absence of accurately identified A. solidipes, some measure of abundance (inoculum), and virulence of Armillaria to allow further refinements of climate-Armillaria root disease predictions. Continued surveys are also needed to further validate the relationship of Armillaria root disease with maladapted forest trees at diverse geographic locations.
Although the ranges of P. menziesii and A. solidipes show considerable overlap, P. menziesii extends further south into Mexico (Oaxaca) and it can exist in different regions with distinct climatic niches where A. solidipes does not occur (Cannon et al., 2008). Any differences in the ranges of P. menziesii and A. solidipes likely explain why the important climate variables (permutation importance > 10%) were different for P. menziesii and A. solidipes models. Mean temperature of warmest quarter (Bio10), precipitation of coldest quarter (Bio19), and precipitation of driest quarter (Bio17) are the significant variables to predict contemporary distribution for P. menziesii; whereas, temperature annual range (Bio7), maximum temperature of warmest month (Bio5), annual mean temperature (Bio1), and minimum temperature of coldest month (Bio6) were the major variables for A. solidipes. Among climate variables affecting P. menziesii distribution, the mean temperature of warmest quarter (Bio10) showed a greatest contribution (35%) followed by the precipitation of coldest (12%) and driest (10.8%) quarters. A previous study (Rehfeldt et al., 2014b) also found that temperature- and precipitation-related variables were important to predict species distribution of P. menziesii.
The climatic variables related to temperature demonstrated significant roles in the spatial distribution of A. solidipes in this study. Because A. solidipes can exhibit an extremely long-term occupancy of a site (e.g., Ferguson et al., 2003), this root pathogen apparently has the capacity of broad tolerance for changing climatic conditions. However, within a site, A. solidipes spreads predominately via vegetative growth of rhizomorphs, and the rate of Armillaria rhizomorph growth varies dependent on climatic factors, but warmer winters could extend the period for rhizomorph growth (e.g., Rishbeth, 1978; Van Der Kamp, 1993; Peet et al., 1996; Cruickshank et al., 1997). In situations where Armillaria spread via basidiospores, higher precipitation and temperatures in autumn will likely favor production of Armillaria basidiocarps, especially in Boreal regions (Ferguson et al., 2003; Heinzelmann et al., 2019). In general, growth and dispersal of Armillaria may specifically benefit from mild and moist seasons, while hot and dry seasons may increase host stress that allows Armillaria to overcome host defenses (Heinzelmann et al., 2019), which suggests that seasonal climates are important factors that contribute to Armillaria root disease.
Although predictions of future suitable climate space for P. menziesii and A. solidipes are dependent on the accuracy of the projected climate grids, the trends indicate that suitable climate space for both species will move northward and toward higher elevations under both SSP2-4.5 and SSP5-8.5 pathways. Under SSP2-4.5 and SSP5-8.5 conditions, while suitable climate for A. solidipes is predicted to decrease in many areas of the inland western United States and BC, Canada, A. solidipes will remain adapted to many areas of this region, where it will likely still persist and likely contribute significantly to decreased forest health and increased mortality where maladapted hosts remain. However, these models do not take adaptation into account, but it is presumed unlikely that either of these relatively long-lived species can dramatically adapt within such a short period time. A plausible hypothesis is that maladaptation (stress) caused by climate change can predispose host trees and result in increased susceptibility to root pathogens (e.g., Kliejunas et al., 2009; Sturrock et al., 2011).
The science-based information to predict potential distribution of Armillaria root disease pathogens and host trees under present and future climate scenarios will inform strategies to reduce Armillaria root disease. For example, predictive models will help identify tree populations at risk and prioritize species and seed source selection to address the objectives of adaptive management. In general, tree species/populations that are predicted to be adapted to future climates can be selected for or planted within sites that are expected to have suitable climate for the Armillaria pathogen. Many existing or naturally regenerated tree populations that exhibit a moderate-to-high degree of phenotypic plasticity for climatic adaptability (i.e., generalists), such as P. monticola, will likely experience less stress from climatic maladaptation and should also exhibit higher resistance/tolerance to Armillaria root disease. Other forest management practices that reduce tree stress, such as increased spacing, may also reduce impacts of Armillaria root disease under a changing climate.
Conclusion
Contemporary and future predictions of suitable climate space of P. menziesii and A. solidipes generated by MaxEnt modeling is an informative tool that can be used to help reduce impacts from Armillaria root disease under changing climates. It is important to note, however, predicted suitable climate space for a species does not necessarily correspond to the future distribution of that species. Although the co-occurrence of suitable climate space for the pathogen and its host does not necessarily result in disease, it is a likely prerequisite for disease. As discussed above, future species distribution and the occurrence of disease will depend on many factors, such as population structure, migration, regeneration, competition, susceptibility to insect attack and diseases, and other interacting factors. The realized distribution of a species will be determined by the interactions among diverse biological, climatic, and environmental factors. Regardless, this study provides an approach to examine the influence of climate on the distribution of an Armillaria root disease pathogen (A. solidipes) and its host tree (P. menziesii). Such approaches are especially useful in determining areas with climate where Armillaria pathogens should not occur, and of course, Armillaria root disease will not occur in areas where the Armillaria pathogen does not occur. The climate-based modeling methods developed from this study can also be used to model other important forest pathogens and examine the potential for invasive species to occupy new geographic areas under contemporary and future climates.
Data Availability Statement
The original contributions presented in the study are included in the article and further inquiries can be directed to the corresponding author.
Author Contributions
NK, JH, JS, MW, GM, and M-SK designed the study and collected a partial Armillaria solidipes data. M-SK, NK, JS, and JH wrote the first draft of the manuscript. JH performed the data analyses. All authors contributed to manuscripts revision and approved the submitted version.
Funding
This project was partially funded by the Forest Health Protection, Special Technology Development Program (R1-2020-2, R4-2019-1, and R2-2018-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank A. L. Smith, H. Maffei, M. L. Fairweather, J. T. Blodgett, P. J. Zambino, J. Worrall, K. S. Burns, J. J. Jacobs, S. M. Ashiglar, J. E. Lundquist, A. L. Ross-Davis, C. Hoffman, R. Mathiasen, R. Hofstetter, J. D. Shaw, E. V. Nelson, M. R. Cleary, S. Brar, B. A. Richardson, and many other collaborators for their help with obtaining Armillaria collections and data.
Footnotes
References
Berbee, M., Bazzicalupo, A., and Le Renard, L. (2014). Direct GenBank Submission KJ146700: fungi of Capilano River Regional Park, North Vancouver BC Canada. Submitted (10-JAN-2014). Vancouver, British Columbia: University of British Columbia.
Berbee, M., Bazzicalupo, A., and Le Renard, L. (2015). Direct GenBank Submission KP454026: fungi of Capilano River Regional Park, North Vancouver BC Canada. Submitted (09-JAN-2015). Vancouver, British Columbia: University of British Columbia.
Blodgett, J. T., Hanna, J. W., Pitman, E. W. I., Ashiglar, S. M., Lundquist, J. E., Kim, M.-S., et al. (2015). “Bioclimatic models estimate areas of suitable habitat for Armillaria spp. in Wyoming” in Proceedings of the 62nd Western International Forest Disease Work Conference. comps M. Murray and P. Palacios (Olympia, WA: Washington Department of Natural Resources). 29–33.
Blodgett, J. T., and Lundquist, J. E. (2011). “Distribution, species, and ecology of Armillaria in Wyoming,” in Proceedings of the 58th Western International Forest Work Conference. comps M. L. Fairweather and P. Palacios (Logan, UT: Utah State University). 58.
Cannon, P., Klopfenstein, N. B., Kim, M.-S., Hanna, J. W., Medel, R., and Alvarado-Rosales, D. (2008). “An Armillaria survey in Mexico: a basis for determining evolutionary relationships, assessing potentially invasive pathogens, evaluating future impacts of climate change, and developing international collaborations in forest pathology” in Proceedings of the 55th Western International Forest Disease Work Conference. comp M. G. Williams (Salem, OR: Oregon Department of Forestry). 29–39.
Chmura, D. J., Anderson, P. D., Howe, G. T., Harrington, C. A., Halofskyd, J. E., Peterson, D. L., et al. (2011). Forest responses to climate change in the northwestern United States: Ecophysiological foundations for adaptive management. For. Ecol. Manag. 261, 1121–1142. doi: 10.1016/j.foreco.2010.12.040
Cruickshank, M. G., and Filipescu, C. N. (2017). The interactive effect of root disease and climate on wood properties in halfsibling Douglas-fir families. For. Ecol. Manag. 392, 58–67. doi: 10.1016/j.foreco.2017.03.002
Cruickshank, M. G., Morrison, D. J., and Lalumière, A. (2011). Site, plot, and individual tree yield reduction of interior Douglas-fir associated with non-lethal infection by Armillaria root disease in southern British Columbia. For. Ecol. Manag. 261, 297–307. doi: 10.1016/j.foreco.2010.10.023
Cruickshank, M. G., Morrison, D. J., and Punja, Z. K. (1997). Incidence of Armillaria species in precommercial thinning stumps and spread of Armillaria ostoyae to adjacent Douglas-fir trees. Can. J. For. Res. 27, 481–490. doi: 10.1139/x96-185
Dorfman, D. D., Berbaum, K. S., and Metz, C. E. (1992). Receiver operating characteristic rating analysis: generalization to the population of readers and patients with the jackknife method. Investig. Radiol. 27, 723–731.
Elías-Román, R. D., Guzmán-Plazola, R. A., Klopfenstein, N. B., Alvarado-Rosales, D., Calderón-Zavala, G., Mora-Aguilera, J. A., et al. (2013). Incidence and phylogenetic analyses of Armillaria spp. associated with root disease in peach orchards in the State of Mexico, Mexico. For. Pathol. 43, 390–401. doi: 10.1111/efp.12043
Eyring, V., Bony, S., Meehl, G. A., Senior, C. A., Stevens, B., Stouffer, R. J., et al. (2016). Overview of the Coupled Model Intercomparison Project Phase 6 (CMIP6) experimental design and organization. Geosci. Model Dev. 9, 1937–1958. doi: 10.5194/gmd-9-1937-2016
Feng, X., Park, D. S., Liang, Y., Pandey, R., and Papeş, M. (2019). Collinearity in ecological niche modeling: confusions and challenges. Ecol. Evol. 9, 10365–10376. doi: 10.1002/ece3.5555
Ferguson, B. A., Dreisbach, T. A., Parks, C. G., Filip, G. M., and Schmitt, C. L. (2003). Coarse-scale population structure of pathogenic Armillaria species in a mixed-conifer forest in the Blue Mountains of northeast Oregon. Can. J. For. Res. 33, 612–623. doi: 10.1139/x03-065
Fick, S. E., and Hijmans, R. J. (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315. doi: 10.1002/joc.5086
Hanna, J. W. (2005). Armillaria ostoyae: genetic characterization and distribution in the western United States. [master thesis]. Moscow, ID: University of Idaho, p. 116.
Hanna, J. W., Blodgett, J. T., Pitman, E. W. I., Ashiglar, S. M., Lundquist, J. E., Kim, M.-S., et al. (2014). “Climate-based species distribution models for Armillaria solidipes in Wyoming: a preliminary assessment” in Proceedings of the 61st Annual Western International Forest Disease Work Conference. comps K. Chadwick and P. Palacios (Logan, UT: Utah State University). 117–120.
Hanna, J. W., Kim, M.-S., Klopfenstein, N. B., Smith, A. L., and Maffei, H. M. (2008). “Determination of suitable climate space for Armillaria ostoyae in the Oregon East Cascades” in Proceedings of the 55th Western International Forest Disease Work Conference. comp M. G. McWilliams (Salem, OR: Oregon Department of Forestry). 92.
Hanna, J. W., Smith, A. L., Maffei, H. M., Kim, M.-S., and Klopfenstein, N. B. (2009). “Survey of Armillaria spp. in the Oregon East Cascades: baseline data for predicting climatic influences on Armillaria root disease,” in Proceedings of the 56th Annual Western International Forest Disease Work Conference. comps F. Baker, C. Jamieson, and P. Palacios (Logan, UT: Utah State University). 53–59.
Hanna, J. W., Klopfenstein, N. B., Kim, M.-S., McDonald, G. I., and Moore, J. A. (2007). Phylogeographic patterns of Armillaria ostoyae in the western United States. For. Pathol. 37, 192–216. doi: 10.1111/j.1439-0329.2007.00497.x
Heinzelmann, R., Dutech, C., Tsykun, T., Labbé, F., Soularue, J.-P., and Prospero, S. (2019). Latest advances and future perspectives in Armillaria research. Can. J. Plant Pathol. 41, 1–23. doi: 10.1080/07060661.2018.1558284
Hermann, R. K., and Lavender, D. P. (1990). “Pseudotsuga menziesii (Mirb.) Franco” in USDA Forest Service, Agriculture Handbook 654. eds R. M. Burns and B. H. Honkala (Washington, DC: U.S. Department of Agriculture). 527–540.
Hertert, H. D., Miller, D. L., and Partridge, A. D. (1975). Interaction of bark beetles (Coleoptera: scolytidae) and root-rot pathogens in grand fir in northern Idaho. Can. Entomol. 107, 899–904.
Hoffman, C. W., Mathiasen, R. L., Hofstetter, R. W., Fairweather, M. L., Shaw, J. D., Hanna, J. W., et al. (2014). Survey for Armillaria by plant associations in northern Arizona. J. Arizona-Nevada Acad. Sci. 45, 76–86. doi: 10.2181/036.045.0204
Johnson, A. L. S., Wallis, G. W., and Foster, R. E. (1972). Impact of root rot and other diseases in young Douglas-fir plantations. For. Chron. 48, 316–319.
Khanum, R., Mumtaz, A., and Kumar, S. (2013). Predicting impacts of climate change on medicinal asclepiads of Pakistan using Maxent modeling. Acta Oecol. 49, 23–31. doi: 10.1016/j.actao.2013.02.007
Kim, M.-S., Klopfenstein, N. B., Hanna, J. W., and McDonald, G. I. (2006). Characterization of North American Armillaria species: genetic relationships determined by ribosomal DNA sequences and AFLP markers. For. Pathol. 36, 145–164. doi: 10.1111/j.1439-0329.2006.00441.x
Kim, M.-S., Klopfenstein, N. B., and McDonald, G. I. (2010). Effects of forest management practices and environment on occurrence of Armillaria species. J. Korean For. Soc. 99, 251–257.
Kim, M.-S., Klopfenstein, N. B., McDonald, G. I., Arumuganathan, K., and Vidaver, A. K. (2000). Characterization of North American Armillaria species by nuclear DNA content and RFLP analysis. Mycologia 92, 874–883. doi: 10.1080/00275514.2000.12061232
Kliejunas, J. T., Geils, B., Glaeser, J. M., Goheen, E. M., Hennon, P., Kim, M.-S., et al. (2009). Climate and forest diseases of western North America: a literature review. Gen. Tech. Rep. PSW-GTR-225. Albany, CA: USDA, Forest Service, Pacific Southwest Research Station, p. 54.
Klopfenstein, N. B., Hanna, J. W., Fairweather, M. L., Shaw, J. D., Mathiasen, R., Hoffman, C., et al. (2012). “Developing a prediction model for Armillaria solidipes in Arizona,” in Proceedings of the 59th Annual Western International Forest Disease Work Conference. comps S. Zeglen and P. Palacios (Logan, UT: Utah State University). 149–152.
Klopfenstein, N. B., Kim, M.-S., Hanna, J. W., Richardson, B. A., and Lundquist, J. E. (2009). Approaches to predicting potential impacts of climate change on forest disease: an example with Armillaria root disease. Res. Pap. RMRS-RP-76. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station, p. 10.
Klopfenstein, N. B., Stewart, J. E., Ota, Y., Hanna, J. W., Richardson, B. A., Ross-Davis, A. L., et al. (2017). Insights into the phylogeny of Northern Hemisphere Armillaria: neighbor-net and Bayesian analyses of translation elongation factor 1-α gene sequences. Mycologia 109, 75–91. doi: 10.1080/00275514.2017.1286572
Kubiak, K., Żółciak, A., Damszel, M., Lech, P., and Sierota, Z. (2017). Armillaria pathogenesis under climate changes. Forests 8:100. doi: 10.3390/f8040100
Kulhavy, D. L., Partridge, A. D., and Stark, A. W. (1984). Root diseases and blister rust associated with bark beetles (Coleoptera: scolytidae) in western white pine in Idaho. Environ. Entomol. 13, 813–817.
Kumar, S., and Stohlgren, T. J. (2009). Maxent modeling for predicting suitable habitat for threatened and endangered tree Canacomyrica monticola in New Caledonia. J. Ecol. Natural Environ. 1, 94–98.
Little, E. L. Jr. (1971). Atlas of United States trees. Volume 1. Conifers and Important Hardwoods. Misc. Publ. no. 1146. Washington, DC: U.S. Department of Agriculture, Forest Service.
Lockman, I. B., and Kearns, H. S. J. (2016). Forest root diseases across the United States. Gen. Tech. Rep. RMRS-GTR-342. Ogden, UT: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station, p. 55.
McDonald, G. I. (1990). “Connecting forest productivity to behavior of soil-borne diseases” in Proceedings-Management and Productivity of Western Montane Forest Soils. comps A. E. Harvey and L. F. Neuenschwander (Ogden, UT: U.S. Department of Agriculture, Forest Service, Intermountain Research Station). 129–144.
McDonald, G. I. (1998). “Preliminary report on the ecology of Armillaria in Utah and the inland west” in Proceedings of the 46th Annual Western International Forest Disease Work Conference. comp L. Trummer (Anchorage, AK: U.S. Department of Agriculture, Forest Service). 85–92.
McDonald, G. I., Hanna, J. W., Smith, A. L., Maffei, H. M., Kim, M.-S., Ross-Davis, A. L., et al. (2011). “Preliminary report on the ecology and predicted suitable climate space of Armillaria in the East Cascades of Oregon” in Proceedings of the 58th Annual Western International Forest Disease Work Conference. comps M. L. Fairweather and P. Palacios (Logan, UT: Utah State University). 135–138.
McDonald, G. I., Harvey, A. E., and Tonn, J. R. (2000). “Fire, competition and forest pests: landscape treatment to sustain ecosystem function” in Proceedings from the Joint Fire Science Conference and Workshop: crossing the Millennium: integrating Spatial Technologies and Ecological Principles for a New Age in Fire Management Volume 2. tech eds L. F. Neuenschwander and K. C. Ryan (Moscow, ID: University of Idaho and the International Association of Wildland Fire). 195–211.
McDonald, G. I., Martin, N. E., and Harvey, A. E. (1987a). Armillaria in the Northern Rockies: pathogenicity and host susceptibility on pristine and disturbed sites. Res. Note INT-371. Ogden, UT: U.S. Department of Agriculture, Forest Service, Intermountain Research Station, p. 5.
McDonald, G. I., Martin, N. E., and Harvey, A. E. (1987b). Occurrence of Armillaria spp. in forests of the Northern Rocky Mountains. Res. Pap. INT-381. Ogden, UT: U.S. Department of Agriculture, Forest Service, Intermountain Research Station, p. 7.
Morrison, D. J. (1981). “Armillaria root disease. A guide to disease diagnosis, development and management in British Columbia” in Information Report. BC-X-203. (Environment Canada: Canadian Forest Service).
Morrison, D. J. (2011). Epidemiology of Armillaria root disease in Douglas-fir plantations in the cedar-hemlock zone of the southern interior of British Columbia. For. Pathol. 41, 31–40. doi: 10.1111/j.1439-0329.2009.00630.x
Morrison, D. J., Pellow, K. W., Norris, D. J., and Nemec, A. F. L. (2000). Visible versus actual incidence of Armillaria root disease in juvenile coniferous stands in the southern interior of British Columbia. Can. J. For. Res. 30, 405–414. doi: 10.1139/x99-222
Murray, M. P., and Leslie, A. (2021). Climate, radial growth, and mortality associated with conifer regeneration infected by root disease (Armillaria ostoyae). For. Chron. 97, 43–51. doi: 10.5558/tfc2021-006
Omdal, D. W., Shaw, C. G. III, Jacobi, W. R., and Wager, T. C. (1995). Variation in pathogenicity and virulence of isolates of Armillaria ostoyae on eight tree species. Plant Dis. 79, 939–944.
Parsons, S., Lewis, K. J., and Psyllakis, J. M. (2003). Relationships between roosting habitat and decay of aspen in the sub-boreal forests of British Columbia. For. Ecol. Manag. 177, 559–570. doi: 10.1016/S0378-1127(02)00448-6
Peet, F. G., Morrison, D. J., and Pellow, K. W. (1996). Rate of spread of Armillaria ostoyae in two Douglas-fir plantation in the southern interior of British Columbia. Can. J. For. Res. 26, 148–151.
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Modell. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Rehfeldt, G. E., Crookston, N. L., Warwell, M. V., and Evans, J. S. (2006). Empirical analysis of plant-climate relationships for the western United States. Int. J. Plant Sci. 167, 1123–1150. doi: 10.1086/507711
Rehfeldt, G. E., Jaquish, B. C., López-Upton, J., Sáenz-Romero, C., St Clair, J. B., Leites, L. P., et al. (2014a). Comparative genetic responses to climate for the varieties of Pinus ponderosa and Pseudotsuga menziesii: realized climate niches. For. Ecol. Manag. 324, 126–137. doi: 10.1016/j.foreco.2014.02.035
Rehfeldt, G. E., Leites, L. P., St Clair, J. B., Jaquish, B. C., Sáenz-Romero, C., López-Upton, J., et al. (2014b). Comparative genetic responses to climate in the varieties of Pinus ponderosa and Pseudotsuga menziesii: clines in growth potential. For. Ecol. Manag. 324, 138–146. doi: 10.1016/j.foreco.2014.02.041
Remya, K., Ramachandran, A., and Jayakumar, S. (2015). Predicting the current and future suitable habitat distribution of Myristica dactyloides Gaertn. using MaxEnt model in the Eastern Ghats, India. Ecol. Eng. 82, 184–188.
Rishbeth, J. (1978). Effects of soil temperature and atmosphere on growth of Armillaria rhizomorphs. Trans. Brit. Mycol. Soc. 79, 213–220.
Robinson, R. M., and Morrison, D. J. (2001). Lesion formation and host response to infection by Armillaria ostoyae in the roots of western larch and Douglas-fir. For. Pathol. 31, 371–385. doi: 10.1046/j.1439-0329.2001.00260.x
Ross-Davis, A. L., Hanna, J. W., Kim, M.-S., and Klopfenstein, N. B. (2012). Advances toward DNA-based identification and phylogeny of North American Armillaria species using elongation factor-1 alpha gene. Mycoscience 53, 161–165. doi: 10.1007/S10267-011-0148-X
Shaw, C. G. III (1989). Armillaria ostoyae associated with mortality of new hosts in Chihuahua, Mexico. Plant Dis. 73, 775.
Steeger, C., and Hitchcock, C. L. (1998). Influence of forest structure and diseases on nest-site selection by sed-breasted nuthatches. J. Wildl. Manag. 62, 1349–1358.
Stewart, J. E., Kim, M.-S., Ota, Y., Sahashi, N., Hanna, J. W., Akiba, M., et al. (2020). Phylogenetic and population genetic analyses reveal three distinct lineages of the invasive brown root-rot pathogen, Phellinus noxius, and bioclimatic modeling predicts differences in associated climate niches. Eur. J. Plant Pathol. 156, 751–766. doi: 10.1007/s10658-019-01926-5
Stewart, J. E., Ross-Davis, A. L., Graça, R. N., Alfenas, A. C., Peever, T. L., Hanna, J. W., et al. (2018). Genetic diversity of Puccinia psidii in the Americas and Hawaii: Potential global implications. For. Pathol. 48:e12378. doi: 10.1111/efp.12378
Sturrock, R. N., Frankel, S. J., Brown, A. V., Hennon, P. E., Kliejunas, J. T., Lewis, K. J., et al. (2011). Climate change and forest diseases. Plant Path. 60, 133–149. doi: 10.1111/j.1365-3059.2010.02406.x
Swart, N. C., Cole, J. N. S., Kharin, V. V., Lazare, M., Scinocca, J. F., Gillett, N. P., et al. (2019). The Canadian Earth System Model version 5 (CanESM5.0.3). Geosci. Model Dev. 12, 4823–4873. doi: 10.5194/gmd-12-4823-2019
Tang, X., Yuan, Y., Li, X., and Zhang, J. (2021). Maximum entropy modeling to predict the impact of climate change on pine wilt disease in China. Front. Plant Sci. 12:652500. doi: 10.3389/fpls.2021.652500
Tkacz, B. M., and Schmitz, R. F. (1986). Association of an Endemic Mountain Pine Beetle Population with Lodgepole Pine Infected by Armillaria Root Rot Disease in Utah. Res. Note INT-353. Ogden, UT: U.S. Department of Agriculture, Forest Service, Intermountain Research Station, p. 7.
Van Der Kamp, B. J. (1993). Rate of spread of Armillaria ostoyae in the central interior of British Columbia. Can. J. For. Res. 23, 1239–1241. doi: 10.1139/x93-156
Van der Star, T., Berbee, M. L., Inderbitzin, P., and Fischer, A. L. (2003). Direct GenBank Submission AY228342: sequencing performed as class project, Biology 323, University of British Columbia. Vancouver, British Columbia: University of British Columbia.
Warwell, M. V., McDonald, G. I., Hanna, J. W., Kim, M.-S., Lalande, B. M., Stewart, J. E., et al. (2019). Armillaria altimontana is associated with healthy western white pine (Pinus monticola) planted in northern Idaho: evidence for in situ biological control of A. solidipes. Forests 10:294. doi: 10.3390/f10040294
Weiskittel, A. R., Crookston, N. L., and Rehfeldt, G. E. (2012). Projected future suitable habitat and productivity of Douglas-fir in western North America. Schweiz. Z. Forstwes. 163, 70–78. doi: 10.3188/szf.2012.0070
Wiley, E. O., Mcnyset, K. M., Peterson, A. T., Robins, C. R., and Stewart, A. M. (2003). Niche modeling perspective on geographic range predictions in the marine environment using a machine-learning algorithm. Oceanography 16, 120–127. doi: 10.5670/oceanog.2003.42
Keywords: climate change, drought, potential distribution, root disease, species distribution modeling, tree maladaptation
Citation: Kim M-S, Hanna JW, Stewart JE, Warwell MV, McDonald GI and Klopfenstein NB (2021) Predicting Present and Future Suitable Climate Spaces (Potential Distributions) for an Armillaria Root Disease Pathogen (Armillaria solidipes) and Its Host, Douglas-fir (Pseudotsuga menziesii), Under Changing Climates. Front. For. Glob. Change 4:740994. doi: 10.3389/ffgc.2021.740994
Received: 14 July 2021; Accepted: 05 November 2021;
Published: 13 December 2021.
Edited by:
Alberto Santini, Institute for Sustainable Plant Protection, National Research Council, Consiglio Nazionale delle Ricerche (CNR), ItalyReviewed by:
Angus J. Carnegie, New South Wales Department of Primary Industries, AustraliaSimone Prospero, Swiss Federal Institute for Forest, Snow and Landscape Research (WSL), Switzerland
Copyright © 2021 Kim, Hanna, Stewart, Warwell, McDonald and Klopfenstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mee-Sook Kim, meesook.kim@usda.gov; Ned B. Klopfenstein, ned.klopfenstein@usda.gov
†These authors have contributed equally to this work and share first authorship
 Mee-Sook Kim
Mee-Sook Kim John W. Hanna2†
John W. Hanna2†  Jane E. Stewart
Jane E. Stewart Marcus V. Warwell
Marcus V. Warwell Ned B. Klopfenstein
Ned B. Klopfenstein