Drosophila melanogaster as a model to study polymicrobial synergy and dysbiosis
- 1Department of Integrative Biomedical & Diagnostic Sciences, Oregon Health and Science University School of Dentistry, Portland, OR, United States
- 2Department of Microbiology, School of Medicine, University of Alabama at Birmingham, Birmingham, AL, United States
The fruit fly Drosophila melanogaster has emerged as a valuable model for investigating human biology, including the role of the microbiome in health and disease. Historically, studies involving the infection of D. melanogaster with single microbial species have yielded critical insights into bacterial colonization and host innate immunity. However, recent evidence has underscored that multiple microbial species can interact in complex ways through physical connections, metabolic cross-feeding, or signaling exchanges, with significant implications for healthy homeostasis and the initiation, progression, and outcomes of disease. As a result, researchers have shifted their focus toward developing more robust and representative in vivo models of co-infection to probe the intricacies of polymicrobial synergy and dysbiosis. This review provides a comprehensive overview of the pioneering work and recent advances in the field, highlighting the utility of Drosophila as an alternative model for studying the multifaceted microbial interactions that occur within the oral cavity and other body sites. We will discuss the factors and mechanisms that drive microbial community dynamics, as well as their impacts on host physiology and immune responses. Furthermore, this review will delve into the emerging evidence that connects oral microbes to systemic conditions in both health and disease. As our understanding of the microbiome continues to evolve, Drosophila offers a powerful and tractable model for unraveling the complex interplay between host and microbes including oral microbes, which has far-reaching implications for human health and the development of targeted therapeutic interventions.
Introduction
Drosophila melanogaster or the “fruit fly”, a simple invertebrate, is becoming an attractive model organism for studying a wide range of topics of complex human biology because it shares significant biological similarity to human systems, and has tractable genetic manipulating tools (Chao et al., 2017). In addition, fly stocks and databases are publicly available and accessible (https://flybase.org/). Importantly, 75% of human genes related to disease have their homologs identified in fruit flies (Yamaguchi and Yoshida, 2018; Cagan et al., 2019), which enables the robust and in-depth study of gene regulation, protein interactions and posttranslational modification of conserved human homologs, and their impact on pathogenesis. Successful transformation of our basic understanding of fruit fly biology to the discovery of new genetic disorders in human disease has promoted the recent expansion of employing Drosophila to model human diseases, which will accelerate the scientific discovery of new signaling and metabolic pathways related to human disease and facilitate the translation of the basic science to clinical applications. The flies are permissive to infection by diverse species of microorganisms that often cause diseases in humans, and as such, is an attractive model for studying bacterial virulence and pathogenesis (Edwards and Kjellerup, 2012; Buchon et al., 2014). Several features of D. melanogaster make it an attractive model to study bacterial interactions, including simple endogenous microbial community, high-throughput screening potential, genetic tractability, cost-effectiveness, and ability to recapitulate key virulence events such as in vitro biofilm formation in microbial colonization of the fly’s crop. The fly crop is found to be the most common anatomic site where bacteria (both stable endogenous species coevolved with flies and temporarily or transiently infected bacteria) reside, which represents an excellent accessible port to explore microbial-microbial interactions as well as the microbial-host interactions. Furthermore, fruit flies are readily adapted to their diets and develop a correlative microbial community, which offers a unique opportunity to determine how nutrients (such as dietary sugars) and other environmental conditions interact with the host genetic components to shape a new polymicrobial landscape that may contribute to host physiology and pathology. In some instances, the development of fly microbiota directly modulates host health and disease (Gould et al., 2018; Brinker et al., 2019), which offers great opportunities to dissect underlying mechanisms.
Polymicrobial interactions of oral microbes in Drosophila melanogaster
Biofilm formation is important for the fitness and virulence of many diverse microbial species especially oral microbes. The Drosophila crop serves as a reservoir for bacterial colonization, and therefore, is used to assess both single and multi-species biofilms. The oral feeding model of D. melanogaster has been used to study microbial interactions that are critical for biofilm formation among various oral microorganisms. One of advantage of using the feeding model to study oral microbial interactions is that the model utilizes sucrose, which is an important substrate that facilitates the attachment of microbes to the tooth surface. It has been well documented that sugary diets disrupt symbiotic microbial community in the oral cavity and promote dysbiosis such as a cariogenic community. Streptococcus mutans forms robust, sucrose-dependent biofilms on the tooth surface, colonizes the oral cavity, and becomes a major contributor of dental caries. Infection of Drosophila with S. mutans using the sucrose feeding protocol reveals that the colonization of S. mutans in the fly depends on a key biofilm matrix enzyme GtfB (Peng et al., 2016b), an important virulence component in humans and in rodent caries model, demonstrating the validity of this model. Like many microbial infections, dental caries is a polymicrobial infection. In addition to contribution by various bacteria, an opportunistic fungal organism Candida albicans is commonly co-isolated with S. mutans from plaque in children with severe early childhood caries, and these two species display synergism in worsening disease progression of dental caries in a rodent model (Bowen et al., 2018). Using the oral feeding model of Drosophila, the polymicrobial synergy between C. albicans and S. mutans is recapitulated. In addition, this model enables the rapid characterization of the specific microbial interaction and identification of microbial virulence factors involved. The S. mutans surface protein antigen I/II, a well-studied adhesin that is not required for single species biofilm formation of S. mutans under sucrose condition, is found to be required for the enhanced colonization of C. albicans in the polymicrobial setting during co-infection of Drosophila melanogaster (Yang et al., 2018) (Table 1). The in vivo results in Drosophila mirror results generated in both in vitro studies and in vivo experiments using a rat model of dental caries, suggesting the Drosophila model can be a reliable tool to examine microbial interactions and uncover new insights into microbial colonization in the oral cavity. Furthermore, the in vitro co-culture under the cariogenic condition (1% sucrose) increases acid production, a hallmark of cariogenic property. The fly model can be readily used to explore this phenotype as the flies gut is known to be acidic, and tolerate acids, and the pH-sensitive dye bromophenol blue can be used to monitor pH changes in the flies’ gut (Iatsenko et al., 2018). We propose this labeling method is feasible in tracking pH changes occurred in the flies’gut, We envision that infection of flies with lactate producing bacteria, such as Lactobacillus gasseri would enable its labeling by bromophenol blue staining. L. casei promotes biofilm formation when co-cultured with S. mutans (Wen et al., 2017). The application of this probe will allow us to determine the synergistic interaction between L. gasseri and S. mutans, or C. albicans and S. mutans in vivo in flies, and evaluate whether antigen I/II is required for the increased acid production during the process of the co-colonization of S. mutans and C. albicans in the flies’ gut as illustrated (Figure 1). Alternatively engineering recombinant microbes with pH-sensitive fluorescent probes could provide an invaluable tool to monitor changes in extracellular pH in real time during co-infection of flies (Tournu et al., 2017). It is well-documented that co-culture of S. mutans and C. albicans synergistically increases acid production in vitro, which can be tracked by the use of a recombinant C. albicans strain tagged with pHluorin2, a pH-sensitive variant of green fluorescent protein engineered by Dr. Palmer’s group (Tournu et al., 2017) as illustrated (Figure 1). The use of the pH-sensitive strain should facilitate noninvasive analysis of multispecies biofilm-associated pH change in vivo and help develop a robust in vivo screen to determine how other oral bacteria interact with C. albicans and manipulate cellular and environmental pH values, a key cariogenic virulence factor (Yang et al., 2018). Future studies will be focused on the development of more robust and versatile probes that are able to report dynamic pH changes during co-infection. In this regard, there are numerous pH-sensitive nanoparticles reported that can be explored in the study of contribution of polymicrobial interactions to pH dynamics (Fulaz et al., 2019; Kromer et al., 2022).
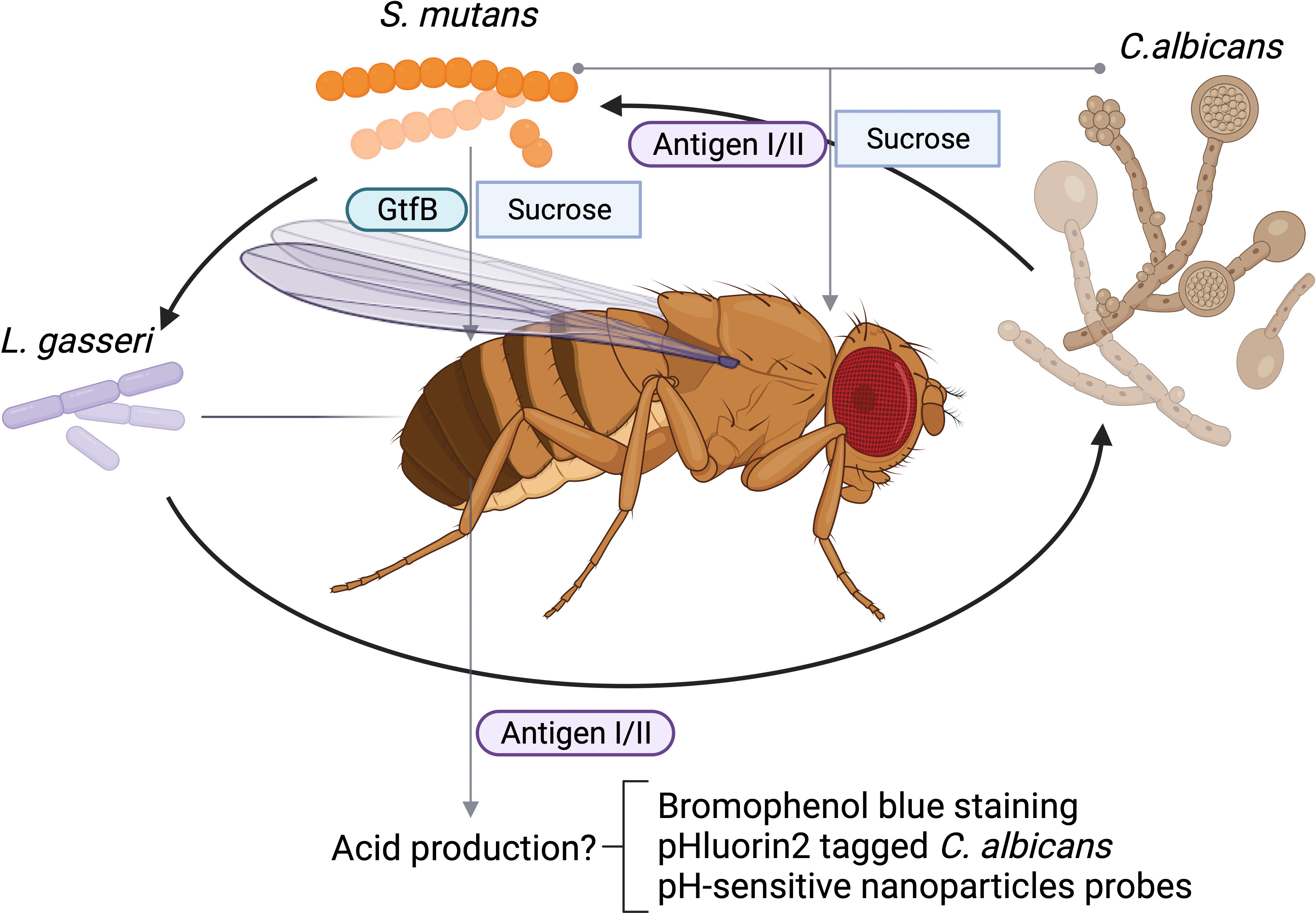
Figure 1 Schematic polymicrobial interactions of oral microbes in D. melanogaster. Acidic condition from L. gasseri fed or Streptococcus mutans and Candida albicans coinfected can be tracked by bromophenol blue staining, an acidic staining, or monitored by fluorescent microscopic examination of labeled C. albicans. C. albicans can be labeled with phlourin2 pH sensor (C. albicans-PHL2). Antigen I/II mediated interactions between C. albicans and and acidic condition can be monitored.
The Drosophila model has also been used to discern interactions between hydrogen peroxide-producing oral commensal Streptococcus parasanguinis and periodontal pathogen Aggregatibacter actinomycetemcomitans. A. actinomycetemcomitans establishes localized aggressive periodontitis in gingival and periodontal pocket sites in adolescents (Fine et al., 2007). Interestingly, the occurrence of S. parasanguinis at subgingival sites with A. actinomycetemcomitans and another oral bacterium is associated with increased bone loss in humans (Fine et al., 2013). Studies evaluating the synergistic relationship between the aforementioned commensal and periopathogen have discovered that A. actinomycetemcomitans promotes the biofilm formation of S. parasanguinis and tightly controls production of hydrogen peroxide by S. parasanguinis. Low concentrations of hydrogen peroxide dose dependently modulate the biofilm formation of S. parasanguinis. These interesting in vitro findings are readily recapitulated in vivo during co-infection of Drosophila with A. actinomycetemcomitans and S. parasanguinis, colonization of S. parasanguinis is significantly increased while the A. actinomycetemcomitans numbers are dramatically inhibited (Duan et al., 2016). Although the underlying mechanisms of inhibition of A. actinomycetemcomitans, and promotion of S. parasanguinis are not entirely clear currently, these results suggest that microbial interactions are more dynamic and complex than previously thought. The fly model provides a powerful tool to investigate complex polymicrobial infections observed in clinic, which should facilitate future in-depth mechanistic studies.
Additionally, oral commensal S. parasanguinis often shares the same ecological niche as Staphylococcus aureus. Using the fly model of colonization, we have determined that S. parasanguinis significantly increased S. aureus biofilm formation and enhanced its colonization in vivo in flies, which highlights how oral commensals may affect the fitness and persistence of S. aureus (Wang et al., 2023). It was determined that streptococcal biofilm-associated protein, BapA1, was essential for dual-species biofilm formation through its colocalization with staphylococcal extracellular DNA, emphasizing the significance of the interspecies biofilm matrix formed between streptococcal BapA1 and staphylococcal eDNA in the formation of polymicrobial interactions. The fly model not only allows mechanistic studies but also provides valuable insights that could inform future research and the development of novel therapeutic approaches targeting polymicrobial biofilms.
Oral streptococci and Pseudomonas aeruginosa interactions in Drosophila melanogaster
Drosophila serves as a natural-route for polymicrobial infection or colonization, and could reflect the synergetic interactions between different species found in in vitro studies or clinical settings (Peters et al., 2012; Gould et al., 2018). Particularly, the current fly feeding model is regarded as a viable platform as it resembles a chronic infection. The localized infection and slow killing kinetics observed following oral infection of Drosophila by P. aeruginosa are consistent with the findings that P. aeruginosa forms micro-colonies or biofilms in the Drosophila crop (Mulcahy et al., 2011). Based on this established single infection model, Drosophila was explored as a tool to screen the interaction of P. aeruginosa with 40 diverse oropharyngeal streptococcal isolates that display varying synergism with P. aeruginosa. High-throughput screens using Drosophila survival as an outcome readout is simple and effective, which enables the identification of specific streptococcal species that potentiate or attenuate P. aeruginosa pathogenesis. Many streptococcal isolates are found to significantly promote the killing of flies by P. aeruginosa, suggesting the interactions between commensal oral streptococci and P. aeruginosa enhance virulence. Such findings are also observed in vivo as the presence of similar organisms in cystic fibrosis (CF) airways associates with the worsened outcome of an existing infection or a newly established infection with P. aeruginosa (Sibley et al., 2008). Intriguingly, opposite effects on virulence by polymicrobial interactions are also documented. Biofilm formation by an oral commensal S. parasanguinis is promoted by P. aeruginosa. P. aeruginosa causes chronic lung infections in CF patients and is a leading contributor of morbidity and mortality in CF disease. Consistently, an oral commensal, S. parasanguinis is commonly found in the CF lung and is associated with improved lung function (Filkins et al., 2012). We have used the Drosophila model to examine whether S. parasanguinis potentially drives positive health outcomes seen in CF lung infections by its interference with the pathogenesis of P. aeruginosa. S. parasanguinis surface adhesins, BapA1 and Fap1, mediate the enhanced colonization of S. parasanguinis, which is dependent on alginate production by P. aeruginosa (Scoffield et al., 2017). The binding of S. parasanguinis to alginate using surface adhesins may explain a potential mechanism of how this bacterium gets incorporated into the CF lung. In fact, another oral streptococcus frequently found colocalized with P. aeruginosa, Streptococcus salivarius employs a carbohydrate binding protein to interface with a different exopolysaccharide of P. aeruginosa to establish its colonization by promoting biofilm formation in vitro and in a D. melanogaster co-infection model(Stoner et al., 2022). Moreover, S. parasanguinis in the presence of nitrite protects Drosophila from killing by P. aeruginosa during co-infections (Scoffield and Wu, 2015), and the nitrite reductase activity by P. aeruginosa is required for the survival of this pathogen during co-infection with S. parasanguinis (Scoffield and Wu, 2016), suggesting a new nitrite-dependent anti-infective mechanism by the oral commensal S. parasanguinis. Interestingly, this mechanism is also relevant to the polymicrobial interaction between commensal S. parasanguinis and a cariogenic bacterium S. mutans. The presence of nitrite enables the hydrogen peroxide producing S. parasanguinis to inhibit S. mutans growth and biofilm formation in vitro. S. parasanguinis effectively competes with S. mutans within the nitrite containing two-species biofilm and inhibits production of a key biofilm matrix, glucans by S. mutans in vitro. These in vitro findings are further validated in an in vivo rat caries model. Commensal S. parasanguinis significantly inhibits cariogenic virulence induced by S. mutans when animals fed nitrite in the drinking water, in comparison with co-infected animals that received no nitrite (Scoffield et al., 2019). These laboratory studies support the clinical observation that high amounts of salivary nitrite are associated with lower prevalence of dental caries (Hohensinn et al., 2016). The protective mechanism offered by commensal streptococci and the host metabolite nitrite provides a potential novel therapeutic strategy by simply modulating dietary nitrite concentrations in the oral cavity.
Pseudomonas aeruginosa and Staphylococcus aureus synergism in Drosophila melanogaster
P. aeruginosa and S. aureus co-infections are common in CF airway disease, suggesting there is a synergistic interaction between two organisms. Indeed, coinfection of animals with P. aeruginosa and S. aureus enhances virulence in two models, a mouse chronic wound model and a rat lung infection model. Such interactions are reproducible in a Drosophila infection model (Korgaonkar et al., 2013). In the oral feeding model, P. aeruginosa kills Drosophila within a few days, and Staphylococcus spp. itself does not kill flies. However, co-infections of Staphylococcus spp with P. aeruginosa increases the killing of flies dramatically while currently decreases the numbers of Staphylococcus spp by 10,000 fold, revealing a synergistic interaction in promoting P. aerugonisa virulence. More interestingly, the coinfection significantly upregulates the expression of genes responsible for Drosophila innate immunity, including various antimicrobial peptides (Sibley et al., 2008). Such synergistic enhancement of innate immunity is not evident using another fly infection model, the septic injury infection, suggesting the oral feeding model is uniquely suitable for the study of co-infection mediated synergy in innate immunity. Mechanistically, P. aeruginosa responds to the major surface component of Staphylococcus aureus, peptidoglycan via a two-component response regulator PA0601, which in turn triggers expression of a host of virulence factors including pyocyanin and elastase. Those virulence factors modulated by the quorum sense can kill both prokaryotic and eukaryotic cells. Importantly, the co-infection enhanced virulence demonstrated in Drosophila oral feeding model recapitulates the outcomes from a murine infection model. The engagement of microbial response regulator in the synergistic interaction is also illustrated in the fly model. In addition, the fly model allows the determination of the requirement of the peptideoglycan signaling in the inhibition of S. aureus growth by P. aeruginosa, which is also readily validated in murine chronic wound model (Korgaonkar et al., 2013). These data further support the notion that Drosophila can be employed as a surrogate host for polymicrobial infections, which offers an effective and efficient alternative to develop new therapeutics targeting polymicrobial interactions, synergistic activation of innate immunity, and numerous polymicrobial cues evident in the model in addition to providing new insights into understanding virulence and pathogenesis of P. aeruginosa in the polymicrobial infections.
Microbial interactions in Drosophila modulate host physiology and pathology
One advantage of using Drosophila as a model is its relative simple gut microbiota (Trinder et al., 2017). The low microbial diversity allows the investigators to systemically determine whether and how microbial function contributes to the development and maturation of host traits (Gould et al., 2018; Brinker et al., 2019). Bacteria metabolize various nutrient ingredients used by flies. Colonization of axenic (microbiota-free) flies with different groups of bacteria regulates various nutrients such as glucose non-specifically. However, modulation of certain nutrients such as triglyceride requires microbial interactions between two major endogenous bacteria, Acetobacter and Lactobacillus (Newell and Douglas, 2014; Sommer and Newell, 2019), highlighting the importance of microbial interactions. It is this simple model that makes comprehensive studies of the association of five major groups of endogenous gut bacteria with fly fitness traits experimentally possible. Emerging studies have revealed how the bacterial interactions support the key function often assigned to a major functional keystone species in terms of their impact on host physiology and pathology such as lifespan and reproduction of flies (Figure 2). The similar results and scenarios have been observed and reported in many other model systems (Chao et al., 2017), suggesting the simple fruit fly model can capture the key events from the complex interactions between host and the microbiome in highly diverse systems such as humans and rodent models. In addition, the recent finding of a symbiotic and persistent strain in the fly gut offers new insights into evolution and adaptation of host-microbe interactions. It is feasible to utilize flies as a model to farm microbes of interests to recapitulate human host-microbe interactions (Gould et al., 2018; Brinker et al., 2019). In vitro bacteria-bacteria interactions can be readily simulated in Drosophila by coinfection, which affords the identification of genetic determinants that are responsible for mutually beneficial or antagonizing interactions, and uncovers a metabolic basis for cross-feeding between bacterial metabolites and their impact on host fitness traits. Microbiota selectively acquired from the natural environment by Drosophila can modulate a wide variety of host cellular pathways and relevant physiology. Microbiota regulate flies’ hematopoietic pathway through a RUNX-like transcription factor, thereby affecting the fly development (Benoit et al., 2017). Recently an elegant study has illustrated the metabolic crosstalk between bacteria and fungi through ethanol catabolism and production of volatile compounds in flies. Metabolic cross feeding by co-culturing of Saccharomyces and Acetobacter triggers differential chemo-sensitive response and enhances egg-laying ability of flies via a conserved olfactory receptor, Or42b (Fischer et al., 2017), revealing a new link between polymicrobe-derived metabolites and host physiology. Olfaction is a crucial element through which flies detect exogenous cues and respond accordingly, which shapes host-microbe interactions including flies’ preference or avoidance for diverse microbes. It is worth noting that such preferences can vary depending on endogenous gut microbiome (Wong et al., 2017), which offers new avenues and opportunities to study dynamic microbiota, genes, and environmental interactions in a simple and tractable system. Another emerging area of studies in microbiome science and medicine is the investigation of brain-gut-microbiome axis in human health and disease (Dinan and Cryan, 2017; Schretter et al., 2018). It has been recognized that the gut has evolved to become a signal hub that integrates a wide range of inputs from different sources including complex microbial and microbe-host interactions. While there are some understanding of bidirectional communications between gut and brain, the underlying molecular mechanisms are poorly defined. The Drosophila is a simple and robust model system to investigate how microbial metabolites from microbial interactions contributes to brain-gut interactions and vice versa, which would offer unique new insights into microbiome-directed gut-brain axis and their contributions to our understanding of the role of microbiota in various neurological disorders such as anxiety, autism, depression or brain tumor. The Drosophila nervous system shares a high degree of similarity with its human counterpart, making it a suitable model for studying the molecular mechanisms underlying development, progression, and potential therapeutic targets of neurological disorders.
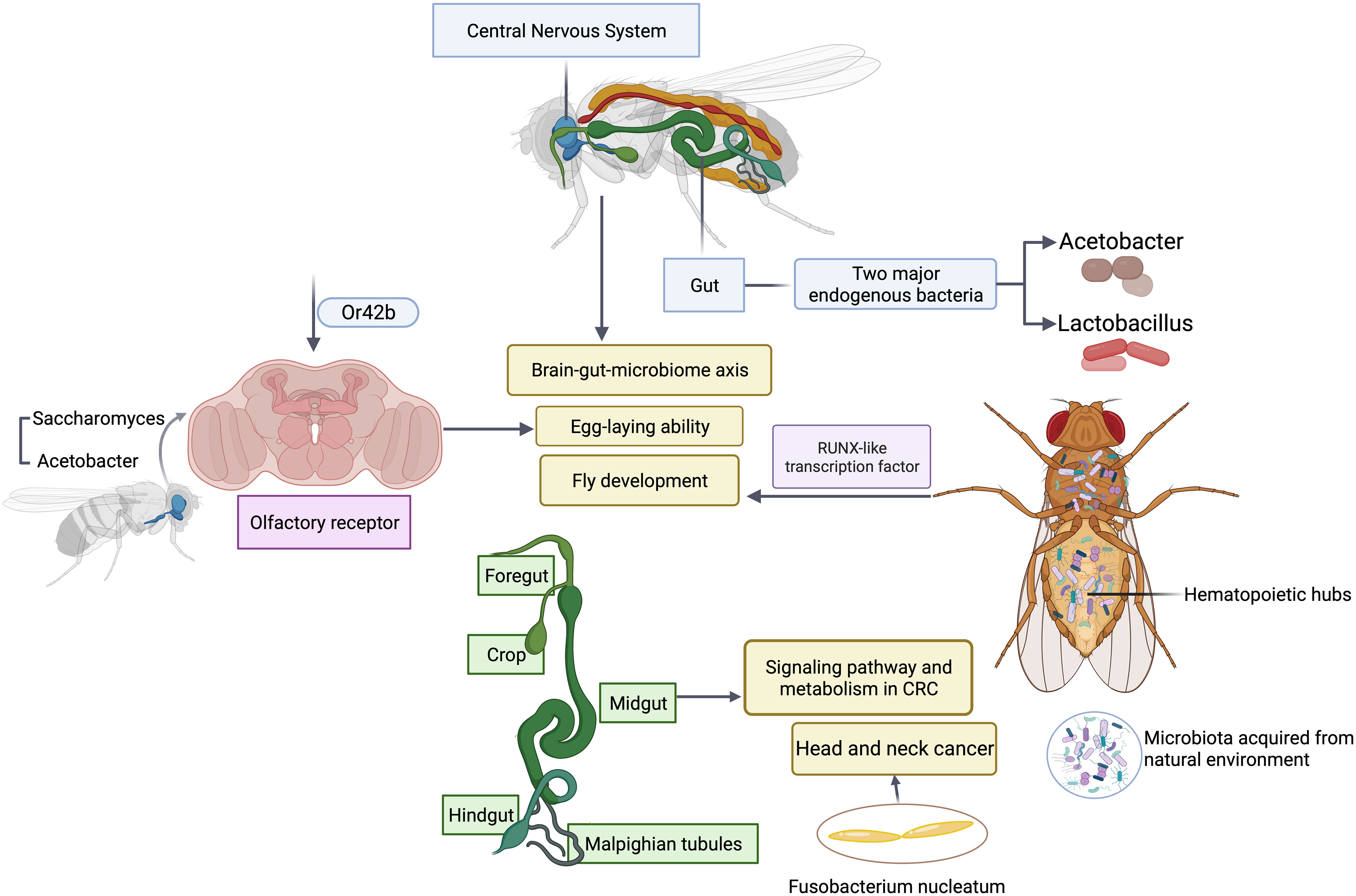
Figure 2 Microbial interactions in Drosophila modulate host physiology and pathology. Polymicrobial infections have a wide range of impacts on gut, central nervous system, olfactory organ and hematopoietic hubs of flies.
In addition, fruit flies can also serve as models for epithelial cancers, including colorectal cancers (CRC). Due to physiological and morphological similarities between fruit flies and mammals, the fly gut, particularly the midgut, is useful for studying the contributions of signaling pathways and metabolism in CRC (Jiang et al., 2022). Recurrent mutations in CRC have been modeled in flies, and multigenic fly models have been developed to unravel the complexity of CRC in terms of metabolism and drug response. These models have provided valuable insights into drug discovery and the functional exploration of human cancer genomes. Furthermore, personalized fly models enable high-throughput screening to develop targeted treatments for patients with refractory metastatic CRC. These studies have demonstrated the practicality of using flies in mechanistic analyses and drug discovery for CRC, highlighting the potential of Drosophila as a useful preclinical whole-animal model.
Given the key role of microbiota played in progression of CRC, notably Fusobacterium nucleatum (Bullman et al., 2017; Clay et al., 2022), it is desirable to employ the fruit flies model to systematically study interplay between microbiota and the development of CRC. This can serve as a feasible model to study the association of microbiota with other types of cancers including head & neck cancer.
The powerful genetic tools offered by the fly model also allows the study of how the residential gut microbiome evolved as a physical and immunological barrier that controls infection of other microbes such as enteric virus. Specific commensals are able to prime inflammatory responses that enables gut epithelial cells to restrict enteric viral infection of flies (Sansone et al., 2015). The fly model enables the rapid identification of various signals mediated by commensal and virus, which provides new perspectives into how dynamic interactions between microbiota and gut epithelial cells direct anti-infective immunity, an emerging exciting area of investigation.
Increasing studies using flies as a model have shown that commensal microbiome affects host aging and lifespan (Guo et al., 2014; Heintz and Mair, 2014; Lee et al., 2019). During the course of aging, the gut bacterial diversity and the overall abundance change substantially (Claesson et al., 2012; Nagpal et al., 2018; Aleman and Valenzano, 2019; Ragonnaud and Biragyn, 2021). Several studies have found these changes affect aging and lifespan in fruit flies (Guo et al., 2014; Clark et al., 2015; Lee et al., 2019). For instance, the shift in microbial composition in aging flies are associated with intestinal barrier failure and shortened lifespan (Clark et al., 2015). Alterations in the microbial composition precede and are linked to dysfunction of the gut epithelial barrier. The intestinal barrier failure in turn leads to a distinct shift in microbiota composition and causes systemic immune activation and the decline in organismal health and lifespan. These results are consistent with findings from an earlier study in which fly mutations that disrupt the septate junctions (SJs) of the gut epithelium induce higher immune activity and concurrently decrease lifespan (Bonnay et al., 2013). Feeding the mutant flies with antibiotics that ablate the commensal bacteria partially restore the lifespan of the mutant flies, showing the link between microbiota and fly lifespan. Additionally, oral infection with bacteria that are able to cross the intestinal barrier, including Serratia marcescens DB11 and Pseudomonas aeruginosa PA14, significantly shorten lifespan of the SJs-disrupted flies, with an increased bacteria load in the hemolymph, indicating the importance of the gut epithelium in modulating microbial infection (Bonnay et al., 2013). Notably, the microbial composition changes are also observed in aged human gastrointestinal tract and in human inflammatory disorder patients (Claesson et al., 2011; Kim and Benayoun, 2020). This similarity further demonstrates Drosophila as one of the viable model organisms to investigate the interplay between microbial interactions, aging and lifespan.
Limitations of the Drosophila model to study polymicrobial interactions
Despite the advantages of the use of the simple, inexpensive and high through-put fly model, it is crucial to recognize the existing limitations of the model. For instance, given the simple microbiota community harbored by flies, sometimes it is challenging to simulate complex microbe-microbe interactions that regulate human health and disease. Thus, careful experimental design is necessary to translate the findings from the D. melanogaster microbiota model (Douglas, 2018). It is well-known that flies lack a comprehensive adaptive immunity system mammals possess (Wong et al., 2016), thus the study of the microbiota and adaptive immunity link is not possible in the model, and some aspects of microbiota-influenced wound healing, tissue repairs and inflammation cannot be modeled. For instance, fibrosis and scarring cannot be readily recapitulated since specific types of cells or tissues responsible for the induction of fibrosis such as myofibroblasts and connective tissues don’t exist in flies. It is also critical to acknowledge the differences in gastrointestinal anatomy such as the absence of lamina propria from the Drosophila intestine, and overall physiology (Lemaitre and Miguel-Aliaga, 2013), which should help guide the design of appropriate studies to address relevant questions in the right context of host-microbe interactions. It is apparent that flies don’t have teeth, while we can mimic bacterial colonization and production of acids, two key events of cariogenesis responsible for the development of dental caries but there is no enamel surface that can be demineralized as evident in rodent animal models (Peng et al., 2016a). Thus, additional complementary models are needed to validate this particular feature in the pathogenesis. By the same token, it is necessary to confirm overall experimental findings from Drosophila using relevant rodent models of infection given the evolutionary divergence between flies and mammals.
Conclusion and future directions
The Drosophila gut microbiome serves as an attractive model of low microbiome complexity. The question remains “Is it possible to establish relatively complex biofilm-like structure formed by polymicrobioal infections which frequently seen in humans and rodents given the simplicity of the endogenous microbial community in flies’ gut?” Another frequently asked question is whether some of the obligate anaerobes such as oral periodontal pathogens P. gingivalis and T. denticola could be incorporated as part of the polymicrobial community because the feeding model relies on the consumption of live bacteria by flies and anaerobes are very sensitive to oxygen killing. This may be circumvented by the use of alternative infection model, a pricking model, which bypasses the gut barriers, enabling inoculation of live bacteria and the study of other unique aspects of microbiome-host interactions (Igboin et al., 2011). In addition, by introducing the capillary feeding system (Murphy et al., 2017), one could constantly feed flies with freshly cultured bacteria and track the amounts of the bacteria the flies consume. It is well documented that dietary conditions, endogenous or invading microbial species, and innate host defense factors collectively determine successful rate of infection of flies by exogenous microbes, which provides the opportunities to establish polymicrobial colonization by taking all those factors into consideration. Such permutations of a wide range of environmental and dietary factors can be readily designed and implemented using the fly model while it would be too expensive to conduct in other systems. In this regard, taking advantage of the known polymicrobial synergy seen among different organisms should help facilitate the colonization of flies with a simple and functional polymicrobial community, in which anaerobes are supported by other microbes to become more resistant to aerobic conditions and then enhance the colonization of the community. This cost-effective, high throughput model makes it possible for the deconvolution of complex systems biology questions. As microbiome has emerged to be an important field in the study of human health and disease, validated fly models of human disease related to cardiovascular, immunological, and metabolic disorders could be readily explored to investigate the functional contributions of microbiota and host-microbe interactions to the development of these systemic conditions. The use of broad spectrum of antibiotics in childhood leads to dysbiosis that is linked to the development of chronic human disorders such as obesity, diabetes and other metabolic disorders, however, the underlying mechanisms are unknown. Epigenetic regulation triggered at the acquisition of dysbiotic microbiota during early life may prime the development of metabolic disorders (Thorburn et al., 2015; Gray et al., 2017), representing a potential microbial inheritable trait. The fly model is a robust system to assess bacterial inheritance, epigenetic regulation, and disease development in offer springs.
In summary, Drosophila is an excellent model for investigating mechanisms of microbe-microbe and host and microbe interactions. The simple microbial diversity makes it possible for developing advanced engineering tools, and techniques to facilitate the study of microbe-fly relationships using systems biology approaches, which should accelerate basic science discoveries and enable potential clinical translations.
Author contributions
HW: conceptualization, data curation, formal analysis, funding acquisition, project administration, resources, software, supervision, validation, visualization, writing – original draft, writing – review & editing. XC: formal analysis, investigation, methodology, validation, writing – original draft, writing – review & editing. JS: conceptualization, formal analysis, writing – original draft, writing – review & editing. BX: investigation, methodology, writing – review & editing. DM: supervision, writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. NIH/NIDCR R01DE017954, R01 DE022350, and NIH/NIDCR R00 DE025913. Funder only provided funding.
Acknowledgments
The work conducted in HW’s laboratory is supported by NIH/NIDCR R01DE017954 and R01 DE022350. JS is supported by NIH/NIDCR R00 DE025913.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aleman, F. D. D., Valenzano, D. R. (2019). Microbiome evolution during host aging. PloS pathogens. 15 (7), e1007727. doi: 10.1371/journal.ppat.1007727
Benoit, J. B., Vigneron, A., Broderick, N. A., Wu, Y., Sun, J. S., Carlson, J. R., et al. (2017). Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. eLife 6. doi: 10.7554/eLife.19535
Bonnay, F., Cohen-Berros, E., Hoffmann, M., Kim, S. Y., Boulianne, G. L., Hoffmann, J. A., et al. (2013). Big bang gene modulates gut immune tolerance in drosophila. Proc. Natl. Acad. Sci. United States America. 110 (8), 2957–2962. doi: 10.1073/pnas.1221910110
Bowen, W. H., Burne, R. A., Wu, H., Koo, H. (2018). Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26 (3), 229–242. doi: 10.1016/j.tim.2017.09.008
Brinker, P., Fontaine, M. C., Beukeboom, L. W., Falcao Salles, J. (2019). Host, symbionts, and the microbiome: The missing tripartite interaction. Trends Microbiol. 27 (6), 480–488. doi: 10.1016/j.tim.2019.02.002
Buchon, N., Silverman, N., Cherry, S. (2014). Immunity in drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14 (12), 796–810. doi: 10.1038/nri3763
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science 358 (6369), 1443–1448. doi: 10.1126/science.aal5240
Cagan, R. L., Zon, L. I., White, R. M. (2019). Modeling cancer with flies and fish. Dev. Cell. 49 (3), 317–324. doi: 10.1016/j.devcel.2019.04.013
Chao, H. T., Liu, L., Bellen, H. J. (2017). Building dialogues between clinical and biomedical research through cross-species collaborations. Semin. Cell Dev. Biol. 70, 49–57. doi: 10.1016/j.semcdb.2017.05.022
Claesson, M. J., Cusack, S., O'Sullivan, O., Greene-Diniz, R., de Weerd, H., Flannery, E., et al. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. United States America. 108 Suppl 1, 4586–4591. doi: 10.1073/pnas.1000097107
Claesson, M. J., Jeffery, I. B., Conde, S., Power, S. E., O'Connor, E. M., Cusack, S., et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488 (7410), 178–184. doi: 10.1038/nature11319
Clark, R. I., Salazar, A., Yamada, R., Fitz-Gibbon, S., Morselli, M., Alcaraz, J., et al. (2015). Distinct shifts in microbiota composition during drosophila aging impair intestinal function and drive mortality. Cell Rep. 12 (10), 1656–1667. doi: 10.1016/j.celrep.2015.08.004
Clay, S. L., Fonseca-Pereira, D., Garrett, W. S. (2022). Colorectal cancer: The facts in the case of the microbiota. J. Clin. Invest. 132 (4). doi: 10.1172/JCI155101
Dinan, T. G., Cryan, J. F. (2017). The microbiome-gut-brain axis in health and disease. Gastroenterol. Clinics North America. 46 (1), 77–89. doi: 10.1016/j.gtc.2016.09.007
Douglas, A. E. (2018). The drosophila model for microbiome research. Lab. animal. 47 (6), 157–164. doi: 10.1038/s41684-018-0065-0
Duan, D., Scoffield, J. A., Zhou, X., Wu, H. (2016). Fine-tuned production of hydrogen peroxide promotes biofilm formation of streptococcus parasanguinis by a pathogenic cohabitant aggregatibacter actinomycetemcomitans. Environ. Microbiol. 18 (11), 4023–4036. doi: 10.1111/1462-2920.13425
Edwards, S., Kjellerup, B. V. (2012). Exploring the applications of invertebrate host-pathogen models for in vivo biofilm infections. FEMS Immunol. Med. Microbiol. 65 (2), 205–214. doi: 10.1111/j.1574-695X.2012.00975.x
Filkins, L. M., Hampton, T. H., Gifford, A. H., Gross, M. J., Hogan, D. A., Sogin, M. L., et al. (2012). Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J. Bacteriol. 194 (17), 4709–4717. doi: 10.1128/JB.00566-12
Fine, D. H., Markowitz, K., Fairlie, K., Tischio-Bereski, D., Ferrendiz, J., Furgang, D., et al. (2013). A consortium of aggregatibacter actinomycetemcomitans, streptococcus parasanguinis, and filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J. Clin. Microbiol. 51 (9), 2850–2861. doi: 10.1128/JCM.00729-13
Fine, D. H., Markowitz, K., Furgang, D., Fairlie, K., Ferrandiz, J., Nasri, C., et al. (2007). Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: Longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45 (12), 3859–3869. doi: 10.1128/JCM.00653-07
Fischer, C. N., Trautman, E. P., Crawford, J. M., Stabb, E. V., Handelsman, J., Broderick, N. A. (2017). Metabolite exchange between microbiome members produces compounds that influence drosophila behavior. eLife 6. doi: 10.7554/eLife.18855
Fulaz, S., Hiebner, D., Barros, C. H. N., Devlin, H., Vitale, S., Quinn, L., et al. (2019). Ratiometric imaging of the in situ ph distribution of biofilms by use of fluorescent mesoporous silica nanosensors. ACS Appl. Mater Interfaces. 11 (36), 32679–32688. doi: 10.1021/acsami.9b09978
Gould, A. L., Zhang, V., Lamberti, L., Jones, E. W., Obadia, B., Korasidis, N., et al. (2018). Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. United States America. 115 (51), E11951–e11960. doi: 10.1073/pnas.1809349115
Gray, L. E., O'Hely, M., Ranganathan, S., Sly, P. D., Vuillermin, P. (2017). The maternal diet, gut bacteria, and bacterial metabolites during pregnancy influence offspring asthma. Front. Immunol. 8, 365. doi: 10.3389/fimmu.2017.00365
Guo, L., Karpac, J., Tran, S. L., Jasper, H. (2014). Pgrp-sc2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 156 (1-2), 109–122. doi: 10.1016/j.cell.2013.12.018
Heintz, C., Mair, W. (2014). You are what you host: Microbiome modulation of the aging process. Cell 156 (3), 408–411. doi: 10.1016/j.cell.2014.01.025
Hohensinn, B., Haselgrubler, R., Muller, U., Stadlbauer, V., Lanzerstorfer, P., Lirk, G., et al. (2016). Sustaining elevated levels of nitrite in the oral cavity through consumption of nitrate-rich beetroot juice in young healthy adults reduces salivary ph. Nitric. Oxide Biol. Chem. 60, 10–15. doi: 10.1016/j.niox.2016.08.006
Iatsenko, I., Boquete, J. P., Lemaitre, B. (2018). Microbiota-derived lactate activates production of reactive oxygen species by the intestinal nadph oxidase nox and shortens drosophila lifespan. Immunity 49 (5), 929–942.e925. doi: 10.1016/j.immuni.2018.09.017
Igboin, C. O., Tordoff, K. P., Moeschberger, M. L., Griffen, A. L., Leys, E. J. (2011). Porphyromonas gingivalis-host interactions in a drosophila melanogaster model. Infect. Immun. 79 (1), 449–458. doi: 10.1128/IAI.00785-10
Jiang, H., Kimura, T., Hai, H., Yamamura, R., Sonoshita, M. (2022). Drosophila as a toolkit to tackle cancer and its metabolism. Front. Oncol. 12, 982751. doi: 10.3389/fonc.2022.982751
Kim, M., Benayoun, B. A. (2020). The microbiome: An emerging key player in aging and longevity. Transl. Med. Aging. 4, 103–116. doi: 10.1016/j.tma.2020.07.004
Korgaonkar, A., Trivedi, U., Rumbaugh, K. P., Whiteley, M. (2013). Community surveillance enhances pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. United States America. 110 (3), 1059–1064. doi: 10.1073/pnas.1214550110
Kromer, C., Schwibbert, K., Gadicherla, A. K., Thiele, D., Nirmalananthan-Budau, N., Laux, P., et al. (2022). Monitoring and imaging ph in biofilms utilizing a fluorescent polymeric nanosensor. Sci. Rep. 12 (1), 9823. doi: 10.1038/s41598-022-13518-1
Lee, H. Y., Lee, S. H., Lee, J. H., Lee, W. J., Min, K. J. (2019). The role of commensal microbes in the lifespan of drosophila melanogaster. Aging (Albany NY). 11 (13), 4611–4640. doi: 10.18632/aging.102073
Lemaitre, B., Miguel-Aliaga, I. (2013). The digestive tract of drosophila melanogaster. Annu. Rev. Genet. 47, 377–404. doi: 10.1146/annurev-genet-111212-133343
Mulcahy, H., Sibley, C. D., Surette, M. G., Lewenza, S. (2011). Drosophila melanogaster as an animal model for the study of pseudomonas aeruginosa biofilm infections in vivo. PloS pathogens. 7 (10), e1002299. doi: 10.1371/journal.ppat.1002299
Murphy, K. R., Park, J. H., Huber, R., Ja, W. W. (2017). Simultaneous measurement of sleep and feeding in individual drosophila. Nat. Protoc. 12 (11), 2355–2366. doi: 10.1038/nprot.2017.096
Nagpal, R., Mainali, R., Ahmadi, S., Wang, S., Singh, R., Kavanagh, K., et al. (2018). Gut microbiome and aging: Physiological and mechanistic insights. Nutr. Healthy Aging. 4 (4), 267–285. doi: 10.3233/NHA-170030
Newell, P. D., Douglas, A. E. (2014). Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in drosophila melanogaster. Appl. Environ. Microbiol. 80 (2), 788–796. doi: 10.1128/AEM.02742-13
Peng, X., Michalek, S., Wu, H. (2016a). Effects of diadenylate cyclase deficiency on synthesis of extracellular polysaccharide matrix of streptococcus mutans revisit. Environ. Microbiol. 18 (11), 3612–3619. doi: 10.1111/1462-2920.13440
Peng, X., Zhang, Y., Bai, G., Zhou, X., Wu, H. (2016b). Cyclic di-amp mediates biofilm formation. Mol. Microbiol. 99 (5), 945–959. doi: 10.1111/mmi.13277
Peters, B. M., Jabra-Rizk, M. A., O'May, G. A., Costerton, J. W., Shirtliff, M. E. (2012). Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25 (1), 193–213. doi: 10.1128/CMR.00013-11
Ragonnaud, E., Biragyn, A. (2021). Gut microbiota as the key controllers of "healthy" aging of elderly people. Immun. Age. 18 (1), 2. doi: 10.1186/s12979-020-00213-w
Sansone, C. L., Cohen, J., Yasunaga, A., Xu, J., Osborn, G., Subramanian, H., et al. (2015). Microbiota-dependent priming of antiviral intestinal immunity in drosophila. Cell Host Microbe 18 (5), 571–581. doi: 10.1016/j.chom.2015.10.010
Schretter, C. E., Vielmetter, J., Bartos, I., Marka, Z., Marka, S., Argade, S., et al. (2018). A gut microbial factor modulates locomotor behaviour in drosophila. Nature 563 (7731), 402–406. doi: 10.1038/s41586-018-0634-9
Scoffield, J. A., Duan, D., Zhu, F., Wu, H. (2017). A commensal streptococcus hijacks a pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PloS pathogens. 13 (4), e1006300. doi: 10.1371/journal.ppat.1006300
Scoffield, J., Michalek, S., Harber, G., Eipers, P., Morrow, C., Wu, H. (2019). Dietary nitrite drives disease outcomes in oral polymicrobial infections. J. Dent. Res. 98 (9), 1020–1026. doi: 10.1177/0022034519855348
Scoffield, J. A., Wu, H. (2015). Oral streptococci and nitrite-mediated interference of pseudomonas aeruginosa. Infect. Immun. 83 (1), 101–107. doi: 10.1128/IAI.02396-14
Scoffield, J. A., Wu, H. (2016). Nitrite reductase is critical for pseudomonas aeruginosa survival during co-infection with the oral commensal streptococcus parasanguinis. Microbiology 162 (2), 376–383. doi: 10.1099/mic.0.000226
Sibley, C. D., Duan, K., Fischer, C., Parkins, M. D., Storey, D. G., Rabin, H. R., et al. (2008). Discerning the complexity of community interactions using a drosophila model of polymicrobial infections. PloS pathogens. 4 (10), e1000184. doi: 10.1371/journal.ppat.1000184
Sommer, A. J., Newell, P. D. (2019). Metabolic basis for mutualism between gut bacteria and its impact on the drosophila melanogaster host. Appl. Environ. Microbiol. 85 (2). doi: 10.1128/AEM.01882-18
Stoner, S. N., Baty, J. J., Scoffield, J. A. (2022). Pseudomonas aeruginosa polysaccharide psl supports airway microbial community development. ISME J. 16 (7), 1730–1739. doi: 10.1038/s41396-022-01221-y
Thorburn, A. N., McKenzie, C. I., Shen, S., Stanley, D., Macia, L., Mason, L. J., et al. (2015). Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 6, 7320. doi: 10.1038/ncomms8320
Tournu, H., Luna-Tapia, A., Peters, B. M., Palmer, G. E. (2017). In vivo indicators of cytoplasmic, vacuolar, and extracellular ph using phluorin2 in candida albicans. mSphere 2 (4). doi: 10.1128/mSphere.00276-17
Trinder, M., Daisley, B. A., Dube, J. S., Reid, G. (2017). Drosophila melanogaster as a high-throughput model for host-microbiota interactions. Front. Microbiol. 8, 751. doi: 10.3389/fmicb.2017.00751
Wang, L., Wang, H., Zhang, H., Wu, H. (2023). Formation of a biofilm matrix network shapes polymicrobial interactions. ISME J. 17 (3), 467–477. doi: 10.1038/s41396-023-01362-8
Wen, Z. T., Liao, S., Bitoun, J. P., De, A., Jorgensen, A., Feng, S., et al. (2017). Streptococcus mutans displays altered stress responses while enhancing biofilm formation by lactobacillus casei in mixed-species consortium. Front. Cell. infect. Microbiol. 7, 524. doi: 10.3389/fcimb.2017.00524
Wong, A. C., Vanhove, A. S., Watnick, P. I. (2016). The interplay between intestinal bacteria and host metabolism in health and disease: Lessons from drosophila melanogaster. Dis. Models mechanisms. 9 (3), 271–281. doi: 10.1242/dmm.023408
Wong, A. C., Wang, Q. P., Morimoto, J., Senior, A. M., Lihoreau, M., Neely, G. G., et al. (2017). Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in drosophila. Curr. Biol. CB. 27 (15), 2397–2404.e2394. doi: 10.1016/j.cub.2017.07.022
Yamaguchi, M., Yoshida, H. (2018). Drosophila as a model organism. Adv. Exp. Med. Biol. 1076, 1–10. doi: 10.1007/978-981-13-0529-0_1
Keywords: bacterial virulence, biofilms, polymicrobial interactions, Drosophila, Streptococcus
Citation: Cao X, Scoffield J, Xie B, Morton DB and Wu H (2023) Drosophila melanogaster as a model to study polymicrobial synergy and dysbiosis. Front. Cell. Infect. Microbiol. 13:1279380. doi: 10.3389/fcimb.2023.1279380
Received: 17 August 2023; Accepted: 01 November 2023;
Published: 21 December 2023.
Edited by:
Ozlem Yilmaz, Medical University of South Carolina, United StatesReviewed by:
Chinmay V. Tikhe, Johns Hopkins University, United StatesCopyright © 2023 Cao, Scoffield, Xie, Morton and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Wu, wuhu@ohsu.edu
 Xixi Cao
Xixi Cao Jessica Scoffield
Jessica Scoffield Baotong Xie1
Baotong Xie1  Hui Wu
Hui Wu