Abstract
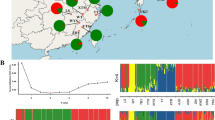
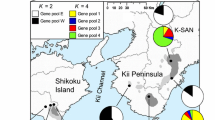
To understand how historical and environmental factors have shaped genetic diversity and spatial genetic patterns of tree species, we characterized the genetic diversity and the genetic structure of a coniferous species, Cryptomeria japonica, which is distributed in Japan (two taxonomic groups) and some parts of southeastern China (C. japonica var. sinensis). We investigated population samples for three groups using high-throughput SNP genotyping, sequencing of multiple genes, and then estimated level of genetic diversity, the genetic differentiation, and the divergence time between the three groups. We also investigated the natural distribution during LGM using species distribution modeling. The genetic variation in the Chinese population, var. sinensis, was lower compared to Japanese populations but it harbored some unique genotypes. The species distribution modeling showed that the Chinese population has decreased since Last Glacial Maximum (LGM) because of lower precipitation than present time, but in Japan the favored environment have remained available for this species even in the LGM. Estimated divergence times of the three groups mostly correspond with the time since the Last Glacial. Our data supports the hypothesis that the glacial period, particularly limitations associated with precipitation, strongly influenced the distribution of C. japonica populations. Our findings suggest that climate change plays an important role in speciation processes of C. japonica.





Similar content being viewed by others
References
Bai WN, Liao WJ, Zhang DY (2010) Nuclear and chloroplast DNA phylogeography reveal two refuge areas with asymmetrical gene flow in a temperate walnut tree from East Asia. New Phytol 188:892–901
Beaumont MA, Zhang W, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162:2025–2035
Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265
Chaney RW (1950) A revision of fossil Sequoia and Taxodium in western North America based on the recent discovery of Metasequoia. Trans Amer Philo Soc 40:171–263
Chen Y, Yang SZ, Zhao MS, Ni BY, Liu L, Chen XY (2008) Demographic genetic structure of Cryptomeria japonica var. sinensis in Tianmushan nature reserve, China. J Integr Plant Biol 50:1171–1177
Cornuet J-M, Santos F, Beaumont MA, Robert CP, Marin J-M, Balding DJ, Guillemaud T, Estoup A (2008) Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24:2713–2719
Cornuet J-M, Ravigné V, Estoup A (2010) Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1. 0). BMC Bioinformatics 11:401
Debreczy Z, Rácz I (2011) Conifer around the world vol.1, DendroPress ltd, Budapest ISBN 978-963-219-063-1
Elith J, Graham C, Anderson R, Dudik M, Ferrier S, Guisan A, Hijmans R, Huettman F, Leathwick J, Lehmann A, Li J, Lohmann L, Loiselle B, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton M, Peterson A, Phillips S, Richardson K, Pereira R, Schapire R, Soberón J, Williams S, Wisz M, Zimmermann N (2006) Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29:129–151
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.). Skeels] endemic to Morocco. Theo Appl Genet 92:832–839
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M (2013) Robust demographic inference from genomic and SNP data. PLoS Genet 9:e1003905
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fischer MC, Rellstab C, Leuzinger M, Roumet M, Gugerli F, Shimizu KK, Holderegger R, Widmer A (2017) Estimating genomic diversity and population differentiation—an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics 18:69
Goodall-Copestake WP, Tarling GA, Murphy EJ (2012) On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals. Heredity 109:50–56
Goudet J (2003) Fstat (ver. 2.9.4), a program to estimate and test population genetics parameters. Available via http://www2.unil.ch/popgen/softwares/fstat.htm
Harrison SP, Yu G, Takahara H, Prentice IC (2001) Palaeovegetation (communications arising): diversity of temperate plants in East Asia. Nature 413:129–130
Hashizume H (1980) Flowering and fructification of Cryptomeria japonica. Iden 34:4–10 (in Japanese)
Hayashi Y (1960) Taxonomical and phytogeographical study of Japanese conifers. Norin-Shuppan, Tokyo
Hayashi R, Takahara H, Inouchi Y, Takemura K, Igarashi Y (2017) Vegetation and endemic tree response to orbital-scale climate changes in the Japanese archipelago during the last glacial-interglacial cycle based on pollen records from Lake Biwa, western Japan. Rev Palaeobot Palynol 241:85–97
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Hewitt GM (1996) Some genetic consequences of ice ages, and their role, in divergence and speciation. Biol J Linn Soc 58:247–276
Howard WR (1997) A warm future in the past. Nature 388:418–419
Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014
Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Igarashi Y, Oba T (2006) Fluctuations in the East Asian monsoon over the last 144 ka in the Northwest Pacific based on a high-resolution pollen analysis of IMAGES core MD01-2421. Quat Sci Rev 25:1447–1459
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kimura M, Kabeya D, Saito T, Moriguchi Y, Uchiyama K, Migita C, Chiba Y, Tsumura Y (2013) Effects of genetic and environmental factors on clonal reproduction in old-growth natural populations of Cryptomeria japonica. For Ecol Manag 304:10–19
Kimura M, Uchiyama K, Nakao K, Moriguchi Y, Jose-Maldia LS, Tsumura Y (2014) Evidence for cryptic northern refugia in the last glacial period of Cryptomeria japonica. Ann Bot 114:1687–1700
Kusumi J, Tsumura Y, Yoshimaru H, Tachida H (2000) Phylogenetic relationships in Taxodiaceae and Cupressaceae sensu stricto based on matK gene, chlL gene, trnL-trnF IGS region, and trnL intron sequences. Am J Bot 87:1480–1488
Kusumi J, Tsumura Y, Tachida H (2015) Evolutionary rate variation in two conifer species, Taxodium distichum (L.) Rich. Var. distichum (baldcypress) and Cryptomeria japonica (Thunb. ex L.f.) D. Don (Sugi, Japanese cedar). Genes & Genetic Systems 90(5):305–315
Leslie AB, Beaulieu JM, Rai HS, Crane PR, Donoghue MJ, Mathews S (2012) Hemisphere-scale differences in conifer evolutionary dynamics. Proc Natl Acad Sci U S A 109:16217–16221
Moriguchi N, Uchiyama K, Miyagi R, Moritsuka E, Takahashi A, Tamura K, Tsumura Y, Teshima K, Tachida H, Kusumi J (2019) Inferring the demographic history of Japanese cedar, Cryptomeria japonica, using amplicon sequencing. Heredity 123:371–383. https://doi.org/10.1038/s41437-019-0198-y
Murai S (1947) Major forestry tree species in the Tohoku region and their varietal problems. In: Kokudo Saiken Zourin Gijutsu Kouenshu, Aomori-rinyukai (eds.) pp.131-151. (in Japanese)
Nagao A, Sasaki S Pharis PR (1989) Cryptomeria japonica. In CRC hand book of flowering. Halevy, AH (ed) vol. VL, CRC press 247-269
Nei M (1977) F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet 41:225–233
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M, Chesser RK (1983) Estimation of fixation indices and gene diversities. Ann Hum Genet 47:253–259
Pakstis AJ, Speed WC, Kidd JR, Kidd KK (2007) Candidate SNPs for a universal individual identification panel. Hum Genet 121:305–317
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research - an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team. R version 2.15.0. (2012) Vienna: R Foundation for Statistical Computing
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19(18):2496–2497
Sakaguchi S, Qiu YX, Liu YH, Qi XS, Kim SH, Han J, Takeuchi Y, Worth JR, Yamasaki M, Sakurai S, Isagi Y (2012) Climate oscillation during the quaternary associated with landscape heterogeneity promoted allopatric lineage divergence of a temperate tree Kalopanax septemlobus (Araliaceae) in East Asia. Mol Ecol 21:3823–3838
Shimizu K (2002) Forestry and Forest products industry in China. The Tropical Forestry 55: 11–20 (in Japanese)
Suzuki E, Susukida J (1989) Age structure and regeneration process of temperate coniferous stands in the Segiri river basin, Yakushima Island. Jpn J Ecol 39:45–51
Taira A (2001) Tectonic evolution of the Japanese island arc system. Annu Rev Earth Planet Sci 29:109–134
Takahara H (1998) Distribution history of Cryptomeria forest. In: Yasuda Y, Miyoushi N (eds) Vegetation history of the Japanese archipelago. Asakura-Shoten, Tokyo, pp 207–223 (in Japanese)
Takahara H, Kitagawa H (2000) Vegetation and climate history since the last interglacial in Kurota lowland, western Japan. Palaeogeogr Palaeoclimatol Palaeoecol 155:123–134
Takahara H, Uemura Y, Danhara T (2000) The vegetation and climate history during the early and mid-last glacial period in Kamiyoshi Basin, Kyoto, Japan. Jpn J Palynology 46:133–146
Tsukada M (1983) Vegetation and climate during the Last Glacial Maximum in Japan. Quat Res 19:212–235
Tsukada M (1986) Altitudinal and latitudinal migration of Cryptomeria japonica for the past 20,000 years in Japan. Quat Res 26:135–152
Tsumura Y, Kado T, Takahashi T, Tani N, Ujino-Ihara T, Iwata H (2007) Genome-scan to detect genetic structure and adaptive genes of natural populations of Cryptomeria japonica. Genetics 176:2393–2403
Tsumura Y, Uchiyama K, Moriguchi Y, Ueno S, Ihara-Ujino T (2012) Genome scanning for detecting adaptive genes along environmental gradients in the Japanese conifer, Cryptomeria japonica. Heredity 109:346–360
Tsumura Y, Uchiyama K, Moriguchi Y, Kimura MK, Ueno S, Ujino-Ihara T (2014) Genetic differentiation and evolutionary adaptation in Cryptomeria japonica. G3 4:2389–2402
Uchiyama K, Ujino-Ihara T, Ueno TY, Futamura N, Shinohara K, Tsumura Y (2012) Single nucleotide polymorphisms in Cryptomeria japonica: their discovery and validation for genome mapping and diversity studies. Tree Genet Genomes 8:1213–1222
Uchiyama K, Miyamoto N, Takahashi M, Watanabe A, Tsumura Y (2014) Population genetic structure and the effect of historical human activity on the genetic variability of Cryptomeria japonica core collection in Japan. Tree Genet Genomes 10:1257–1270
Wang W-T, Xu B, Zhang D-Y, Bai W-N (2016) Phylogeography of postglacial range expansion in Juglans mandshurica (Juglandaceae) reveals no evidence of bottleneck, loss of genetic diversity, or isolation by distance in the leading-edge populations. Mol Phylogenet Evol 102:255–264
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7(2):256–276
Wright S (1922) Coefficients of inbreeding and relationship. Am Nat 56:330–338
Wright S (1978) Evolution and the genetics of populations. Variability within and among natural populations, vol 4. The University of Chicago Press, Chicago
Qi X-S, Chen C, Comes HP, Sakaguchi S, Liu Y-H, Tanaka N, Sakio H, Qiu Y-X (2012) Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian tertiary relict (Cercidiphyllaceae). New Phytol 196(2):617–630
Qiu Y-X, Sun Y, Zhang X-P, Lee J, Cheng-Xin F, Comes HP (2009) Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to quaternary climate change and landbridge configurations. New Phytol 183(2):480–495
Yamazaki T (1995) Cryptomeriaceae. Pp.264, Iwatsuki K, Yamazaki T, Boufford DE, Ohba H (eds) Flora of Japan. Volume I, Pteridophyta and Gymnospermae. Kodansha, Tokyo
Yasue M, Ogiyama K, Suto S, Tsukahara H, Miyahara F, Ohba K (1987) Geographical differentiation of natural Cryptomeria stands analyzed by diterpene hydrocarbon constituents of individual trees. J Jpn For Soc 69:152–156
Yoshida S, Imanaga M (1990) The stand structure anf the growth of sugi (Cryptomeria japonica D. DON) natural forests on Yakushima. J Jpn For Soc 72:131–138
Acknowledgments
We thank Y. Taguchi and M. Koshiba for technical assistance for DNA analysis. This study was partly supported by the Program for the Promotion of Basic and Applied Research for Innovations in Bio-oriented Industry and JSPS KAKENHI program (grant nos. 26292087, 26291082 and 15H02833), the National Key Research and Development Program of China (grant no. 2016YFE0127200), and Environment Research grant of the Sumitomo Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Gugerli
Data archive
All SNPs information and sequence data were deposited in DDBJ (accession numbers, AB874696-AB889383 and LC477701-LC479084).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tsumura, Y., Kimura, M., Nakao, K. et al. Effects of the last glacial period on genetic diversity and genetic differentiation in Cryptomeria japonica in East Asia. Tree Genetics & Genomes 16, 19 (2020). https://doi.org/10.1007/s11295-019-1411-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-019-1411-0