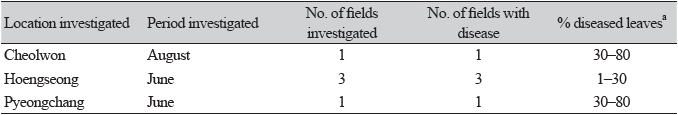
Beet (Beta vulgaris subsp. vulgaris L.), belonging to the family Amaranthaceae, is a biennial or perennial plant native to Azores, western Europe to the Mediterranean region, and India [1]. This plant is widely cultivated as a vegetable in various regions of Korea. From June to August 2021, we surveyed diseases affecting beet plants grown in Cheolwon, Hoengseong, and Pyeongchang regions in Gangwon Province, Korea. We observed severe leaf spot symptoms, such as brown to dark circular or irregular spots on the leaves, in plants (Fig. 1A and B). Three sites in each field and 100 leaves at each site were investigated to determine the disease incidence. Disease incidence on the leaves in the fields investigated at the three locations ranged from 1 to 80% (Table 1).
Leaves of beet plants with spot symptoms were collected from the investigated fields, and fungal isolates were obtained from diseased leaves, as previously described [2]. Most isolates were identified as Phoma sp. based on their morphological characteristics as well as those described by Boerema et al. [3]. Five single-spore isolates were obtained from the original isolates and used for further identification and pathogenicity tests.
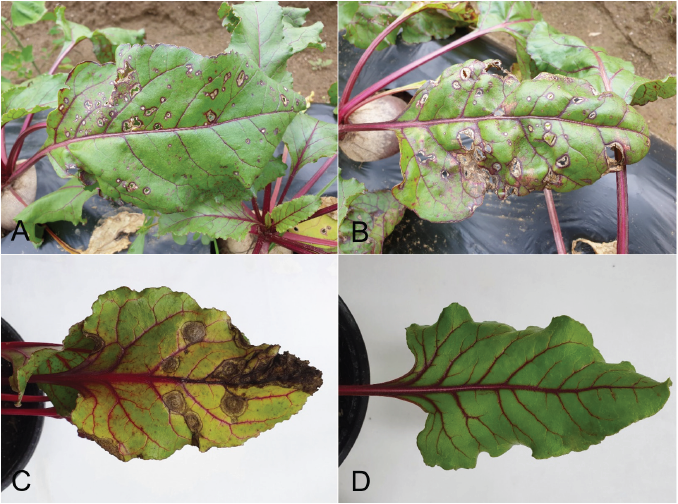
To determine their cultural and morphological characteristics, all single-spore isolates were cultured on malt extract agar (MEA), oatmeal agar (OA), and potato dextrose agar (PDA) at 22℃ for 14 days using the previously described methods [3, 4]. Average diameters of the 7-day-old colonies of the isolates grown on MEA, OA, and PDA in the dark were 5.3, 6.7, and 6.4 cm, respectively. Colonies on MEA exhibited slightly irregular margins and olivaceous to dark olivaceous mycelia (Fig. 2A). Colonies on OA exhibited regular margins and olivaceous to grayish dark olivaceous mycelia (Fig. 2B). Colonies on PDA exhibited olivaceous to grayish olivaceous mycelia (Fig. 2C). NaOH spot test [3] results on 7-day-old MEA cultures of the isolates were negative.
Morphological characteristics of each isolate were determined using 30 pycnidia and 30 conidia from 14-day-old cultures on OA. Pycnidia were variable in size and shape, dark brown to black in color, mostly globose to subglobose, solitary or confluent, semi-immerged in media, from which a creamy conidial mass leaked (Fig. 2D and E), and measured 170‒392 μm (av. 254 μm) in diameter. Conidia were aseptate, oblong to ellipsoidal (Fig. 2F), and measured 3.6‒5.9 × 2.0‒4.0 μm (av. 4.7 × 2.9 μm).
Cultural and morphological characteristics of all isolates were similar to those of Phoma betae A.B. Frank. [teleomorph: Pleospora betae (Berl.) Nevod.], as previous described [3]. However, a phylogenetic study reclassified P. betae as Neocamarosporium betae (Berl.) Ariyaw. and K.D. Hyde [5]. To determine the molecular characteristics of the isolates, partial large subunit nuclear ribosomal DNA (LSU) and RNA polymerase II (RPB2) regions of the isolates were investigated using LR0R [6] and LR7 [7], and RPB2-5f2 [8] and fRPB2-7cR [9], respectively. Genomic DNA of the isolates was extracted using the protocol described by Park et al. [10], with slight modifications. All polymerase chain reaction experiments were conducted as previously described [4]. Genomic DNA was sequenced using the primer sets from Bionics (Seoul, Korea). A phylogenetic tree was constructed using the neighbor-joining method with a maximum composite likelihood model including 1,000 bootstrap replicates using MEGA version 7 software [11]. Final concatenated alignments included four ingroup taxa with 1,866 characters containing gaps (1,268 for LSU and 598 for RPB2). Coniothyrium palmarum (CBS 400.71) was set as the outgroup taxon. The phylogenetic tree revealed that all isolates were clustered with the two strains (CBS 109410 and CBS 523.66) of N. betae (Fig. 3). Sequence data of LSU and RPB2 genes obtained from the five isolates were deposited in GenBank with accession numbers of OP077314‒OP077318 and OP094074‒OP094078, respectively.
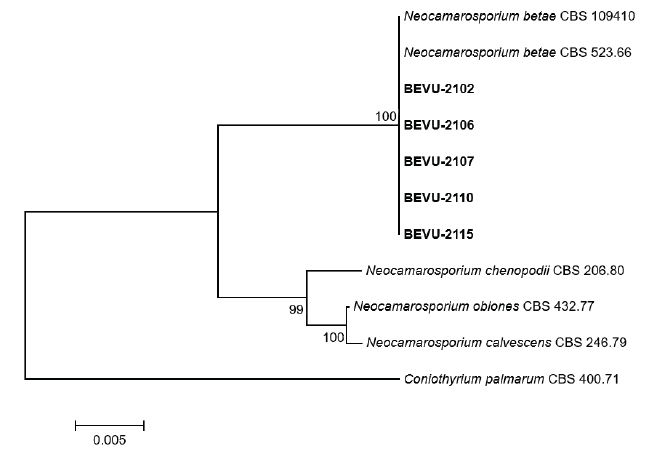
Fig. 3. Phylogenetic tree based on the concatenated sequences of partial large subunit nuclear ribosomal DNA and RNA polymerase II of the five Neocamarosporium betae isolates (BEVU-numbers) from beet and Neocamarosporium spp. Sequence data of Neocamarosporium spp. were obtained from the National Center for Biotechnology Information GenBank database. A phylogenetic tree was generated using the neighbor-joining method with a maximum composite likelihood model including 1,000 bootstrap replicates using MEGA version 7 software. Bootstrap support values are given at the nodes. Bar indicates the number of nucleotide substitutions per site. Tree was rooted to Coniothyrium palmarum CBS 400.71.
Three isolates of N. betae were tested to confirm their pathogenicity in beet plants via artificial inoculation. To produce conidia, isolates were cultured on PDA at 22℃ in the dark for three weeks, after which they were incubated under alternating cycles of 13/11 hr NUV light/dark for two weeks. Subsequently, a conidial suspension (1‒2 × 106 conidia/mL) of each isolate was prepared from 5-week-old cultures, and 20 mL of the conidial suspension of each isolate was sprayed onto every 73-day-old beet plant grown in a plastic pot (height: 13.5 cm; upper diameter: 15 cm; lower diameter: 9 cm) in a vinyl greenhouse. A control plant was treated with 20 mL of sterile distilled water. All inoculated plants were placed in plastic boxes (71.0 × 53.5 × 40.5 cm) under 100% relative humidity at room temperature (24‒26℃) for three days, removed from the the boxes, and grown in a vinyl greenhouse. Then, pathogenicity of the isolates was determined based on the degree of leaf spot symptoms seven days after inoculation. An inoculation test was performed in triplicate. All tested isolates were found to cause leaf spot symptoms in the inoculated plants (Fig. 1C), whereas no symptoms were detected in the control plants (Fig. 1D). The symptoms in inoculated plants were similar to those observed in the plant leaves in the investigated fields. Re-isolation of the inoculated isolates from the lesions further confirmed this result.
N. betae (synonym: P. betae) causes black leg, damping-off, root rot, and leaf spot in beet (3). It also causes leaf spot of spinach in Spain (12). Beet leaf spot caused by N. betae has been reported in New Zealand, USA, and Italy [13, 14, 15]. Although beet leaf spot caused by P. betae is recorded in Korea [16], the identity and pathogenicity of the causal pathogen remain unknown. In this study, we identified N. betae as the pathogen causing beet leaf spot in Korea.